Hello Everyone,
We have recently generated two de-novo transcriptomics assembly for two different but related species. These new transcripts seem quite good on the basis of quality measurements, completeness and alignment with the previously sequenced genome and annotation. We were able to pick up novel genes and previously unannotated transcripts. In order to pick the alternate spliced transcripts we are currently trying running PASA (Program to Assemble Spliced Alignment) annotation.
After PASA annotation, when I made a comparison between trinity transcripts, already existing annotation and PASA annotation in IGB. I find out a case where trinity transcripts fully supported by previously curated annotation as well as the RNASeq data, but no PASA annotation. PASA annotation only shows one fragment, rest of the parts not even present in the valid and failed .gff3 file generated by PASA [Figure]. This leads to few questions - for which I don't have any answer. Here are my questions:
- Why there is this difference in the PASA annotation- when there is already manual curation and RNAseq depth evidences are present? And which one is correct?
- PASA annotation using blat and gmap for the alignment of transcripts to the genome. And we have also used blat to align the transcript to the genome. Then why there is different in two blat alignment?
Data used for Transcriptomics assembly = 100 bp Paired end reads, non-strand specific
Attached figure description:-
- Dark Blue colour = New assembled transcript annotation [this is the annotation generated by aligning assembled transcripts with the genome using blat].
- Orange = existing curated annotation.
- Red = PASA annotation
- Blue colours = shows valid alignment annotation for blat and gmap respectively.
- Read Depth = Green for control and Dark red = Knock-out
Figure: 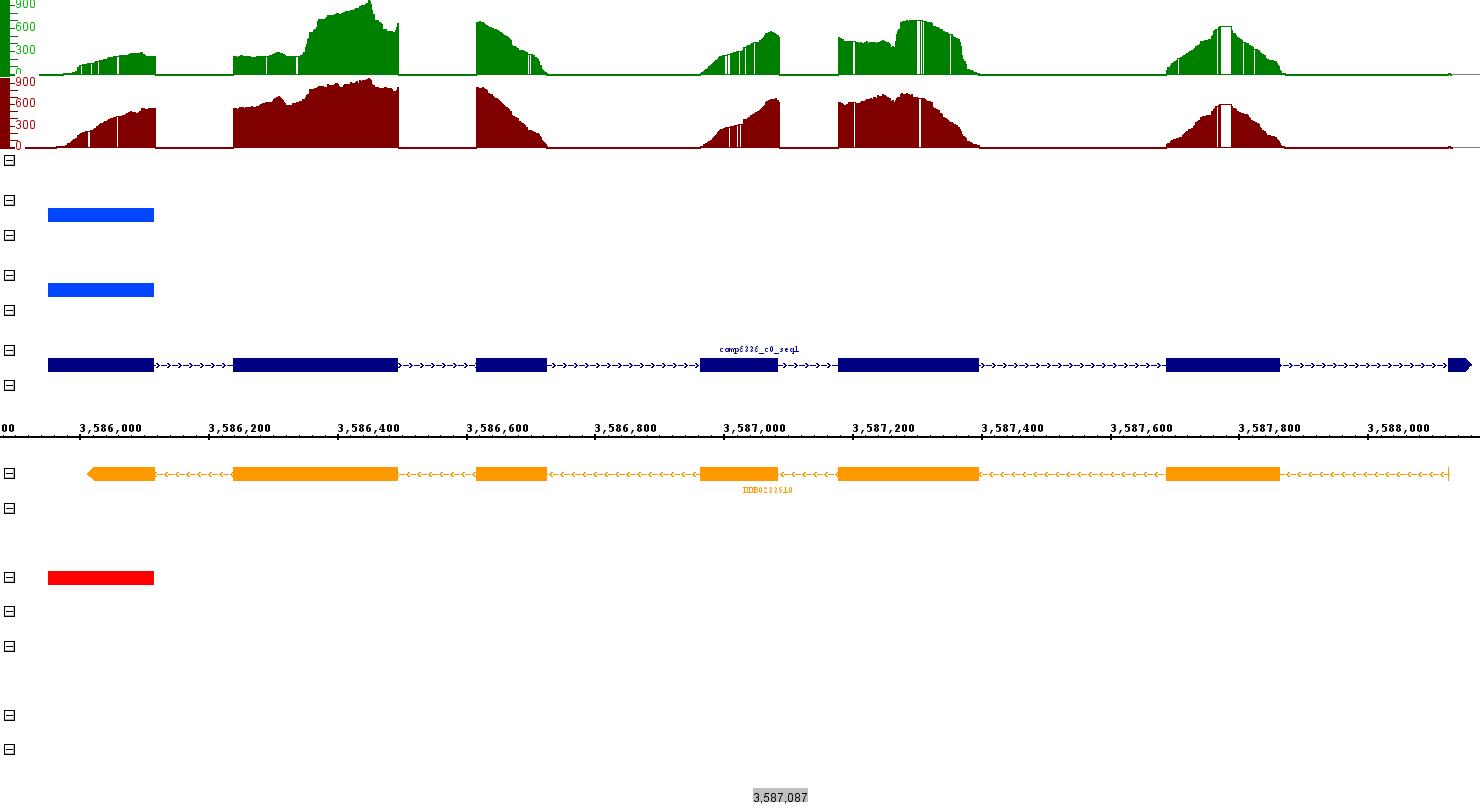
I am very much looking forward for the reply. Any suggestions/view would be very helpful.
Many Thanks
Reema,
PS = I also posting the same post in the seq-answer.


Ok, The problem solved. It seems like the problem associated with .gff3 file. PASA annotation generates three annotation file .gff3,.gtf and .bed. Instead of .gff3 file if I upload .gtf file in IGB then it shows the full transcript annotation. However, there is still some transcripts bits missed in .gtf. But .bed format works great on both .gtf and .gff3.
Hi Reema,
I troubleshoot issues for IGB, and I would like to know more about what's happening with your files.
In order for me to get to the bottom of this, can you please provide the .gff3 and the .bed file?
Thanks!
Nowlan
Hello Nowlan
Could you please send me the ID where I can send you these files?
Best,
Reema,
Hello Reema,
My University email is nfreese@uncc.edu, and I also have a gmail account: nowlan.freese@gmail.com. Either should work, if the files are large they might need to be sent to my gmail account via Google Drive.
Thanks!
Nowlan