I have called variants for our RNA-Seq data following the best practice as mentioned. I am unable to interpret some of the entries in the vcf files. I am pasting the examples below.
chr1 1288459 . TG T 283.73 . AC=1;AF=0.500;AN=2;BaseQRankSum=-2.064;ClippingRankSum=-0.268;DP=50;FS=0.000;MLEAC=1;MLEAF=0.500;MQ=35.71;MQ0=0;MQRankSum=0.268;QD=5.67;ReadPosRankSum=-0.755 GT:AD:DP:GQ:PL 0/1:25,23:48:99:321,0,344
Here, it states that the variant is heterozygous with the DP 48 and ref count AD 25 and alt count AD 23. Quality looks good and the confidence GQ is also very high.
Why are two nucleotides mentioned in REF column? How is it heterozygous if its T in REF and T in ALT columns?
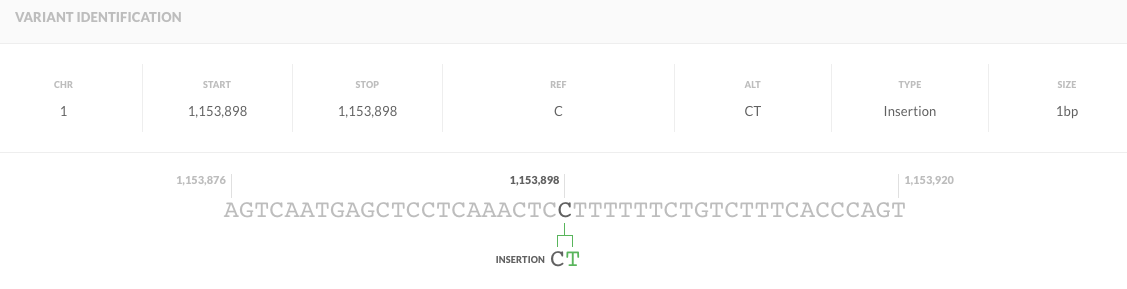
I also find the similar entries with more than two nucleotides in the REF column.
chr1 160209736 . AAGG A 591.52 . AC=1;AF=0.500;AN=2;BaseQRankSum=-0.804;ClippingRankSum=-0.291;DP=44;FS=0.000;MLEAC=1;MLEAF=0.500;MQ=36.53;MQ0=0;MQRankSum=-0.222;QD=13.44;ReadPosRankSum=3.505 GT:AD:DP:GQ:PL 0/1:6,38:44:7:628,0,7
Lastly, the same behavior is also observed the other way around.
chr1 1153898 . C CT 66.73 . AC=1;AF=0.500;AN=2;BaseQRankSum=-1.787;ClippingRankSum=-0.208;DP=77;FS=0.000;MLEAC=1;MLEAF=0.500;MQ=45.50;MQ0=0;MQRankSum=1.633;QD=0.87;ReadPosRankSum=-0.659 GT:AD:DP:GQ:PL 0/1:50,23:73:99:104,0,547
Can anyone shed some light on such entries in VCF?

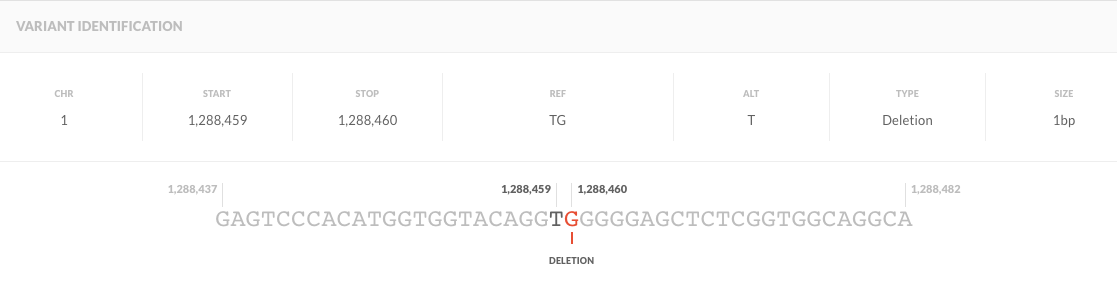

"Why are two nucleotides mentioned in REF column? " because that's a deletion of a G after the T at position chr1:1288459