Entering edit mode
8.8 years ago
rna-seq_researcher
▴
60
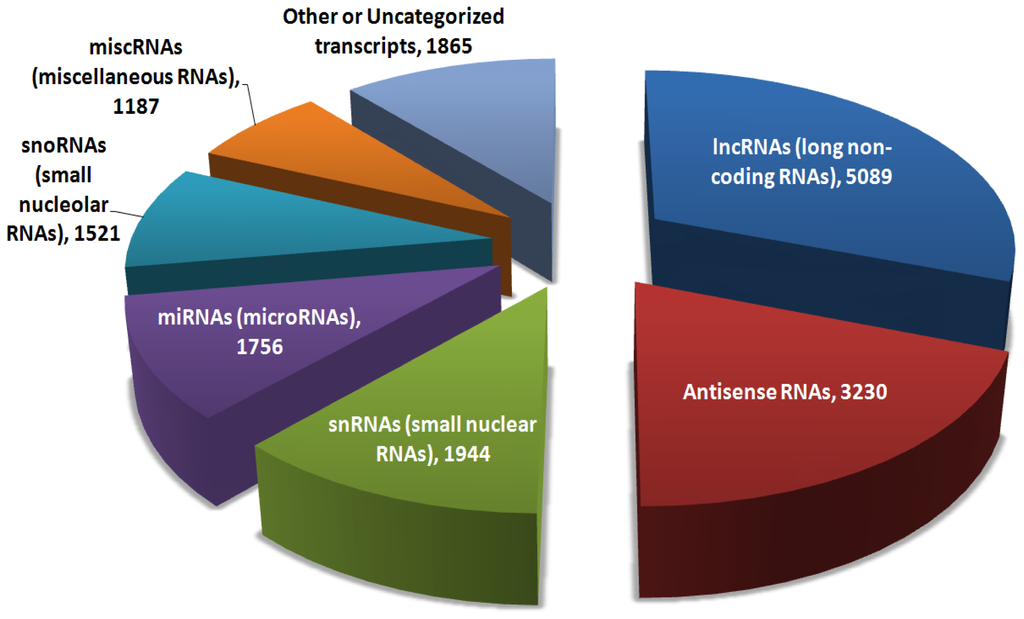
Hi all, I'm looking for a tool I can use to categorize the RNA's from my seq experiment similarly to what is shown below. I saw somewhere that Picard tools may do the trick, but I couldn't find this functionality after a quick look.
Any suggestion would be greatly appreaciated :)