Entering edit mode
8.5 years ago
reza.jabal
▴
580
Hello Guys,
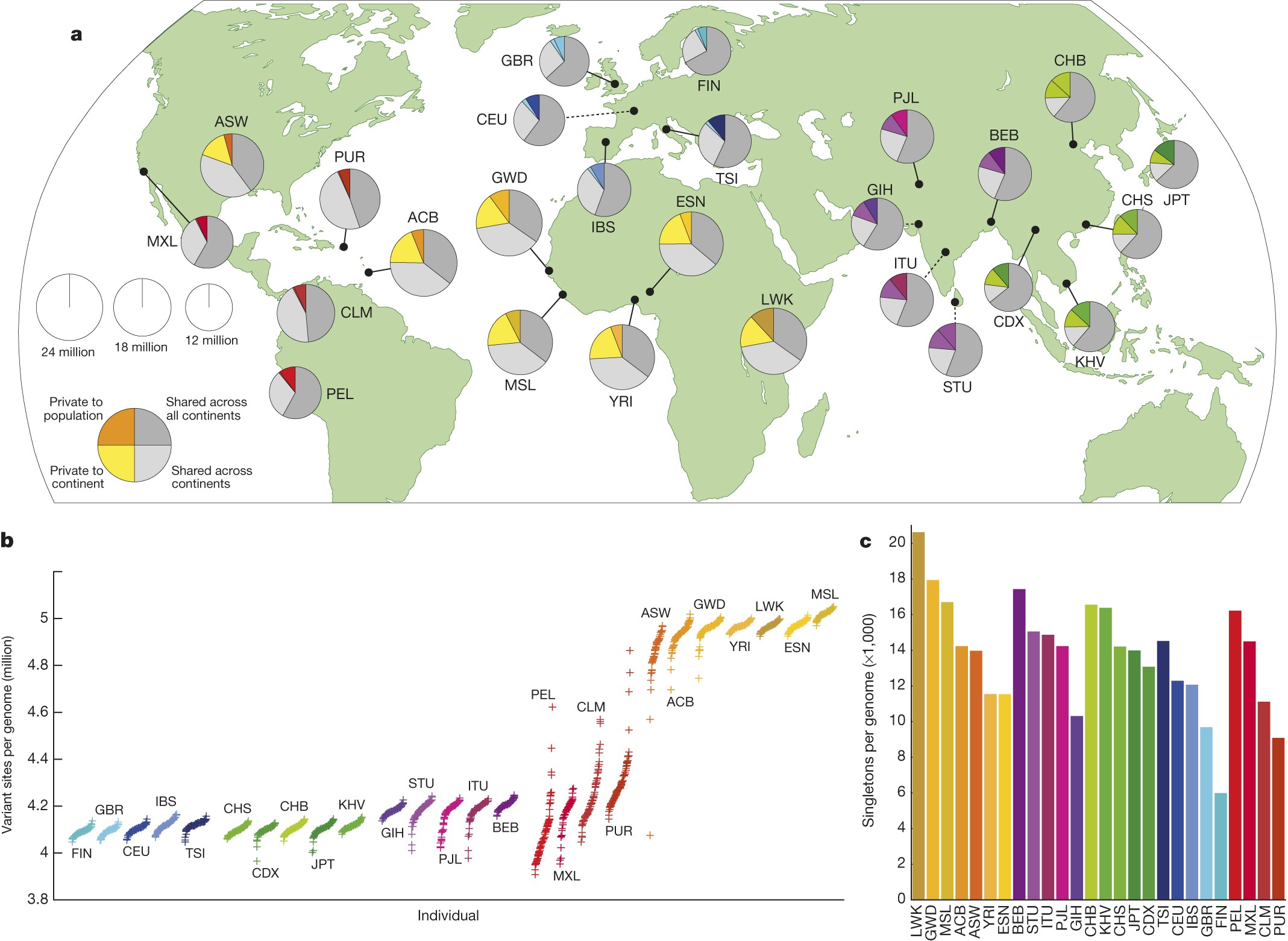
The 1000 Genomes consortium published a paper just couple of days ago on conclusion of the project in nature. According to them a typical genome differs from the reference human genome at 4.1 million to 5 million sites and along with that they produced a nice representation of variants frequency per populations.
As it is expected African populations show greatest number of variants in comparison to other populations, but I am wondering why in the "Exome Variant Server" number of variants for almost every sites is underrepresented for African-American population?


Can you re-phrase your question? I'm interested, but don't quite understand what you are trying to resolve.
Sorry for badly written question. I am simply wondering why for each individual gene in EVS, number of variants that are indexed for African-American population is less than European-American population?
Consistent with "out of Africa model of human origins", we would expect greater number of variants per site in African descendant individuals.