Entering edit mode
7.9 years ago
sosal
▴
10
Hi all.
I have been trying to use ExomeCNV to make an input for EXPANDS (tumor purity tool) (EXPANDS journal paper says "Copy-number segments were calculated using an approach similar to ExomeCNV")
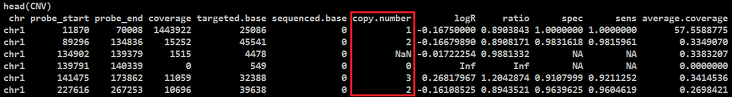
This is CNV input format of EXPANDs
chr startpos endpos CN_Estimate
22 26290824 51237893 1.280513
17 54279 81188538 1.723031
8 162897 146279898 1.865630
5 98191824 110460104 1.740130
14 19114214 107283536 2.016270
15 20087214 56658198 2.011064
But ExomeCNV always makes an integer copy number (CN) results. like this:
I used ExomeCNV functions with default parameters in R.
logR = calculate.logR(normal, tumor)
eCNV = classify.eCNV(normal=normal, tumor=tumor, logR=logR)
CNV = multi.CNV.analyze(normal, tumor, logR=logR, all.cnv.ls=list(eCNV))
My question: I want to make an floating point CN with ExomeCNV, like EXPANDS input format. What should I do?