In a transcript file (.gtf) we can able to get exon wise start position and stop position, CDS and start codon region and stop codon region,but I want start position and end position of all genes (not exon wise) with its chromosome number in an organism.
Where to get Gene start position to end positon for all gene in an organism
0
Entering edit mode
7.9 years ago
Bioblazer
▴
50
3
Entering edit mode
7.9 years ago
Devon Ryan
104k
Normally there's a "gene" entry for each gene, so:
awk 'BEGIN{FS="\t"; OFS="\t"}{if($3 == "gene") print $1, $4, $5}' foo.gtf
2
Entering edit mode
7.9 years ago
EagleEye
7.5k
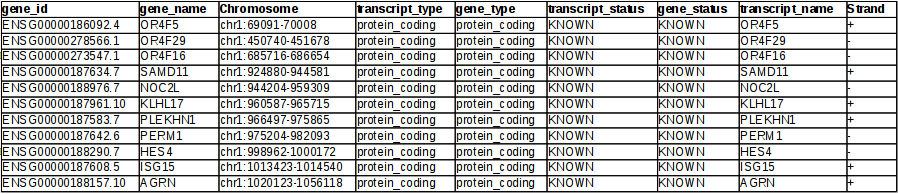
As Devon said if the GTF file is from Gencode, you will have "gene" entry. Extracting "gene" entry will give you the desired results. If you use Ensembl GTF annotation (hg19 upto GRCh37.74), it does not have "gene" entry. In this case use the following script.
The output will look similar to
Similar Posts
Loading Similar Posts
Traffic: 3209 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Thank you very much for your kind reply