Dear Bioinformaticians,
I was looking at various Illumina sequencing platforms here http://www.illumina.com/systems/sequencing-platforms.html.
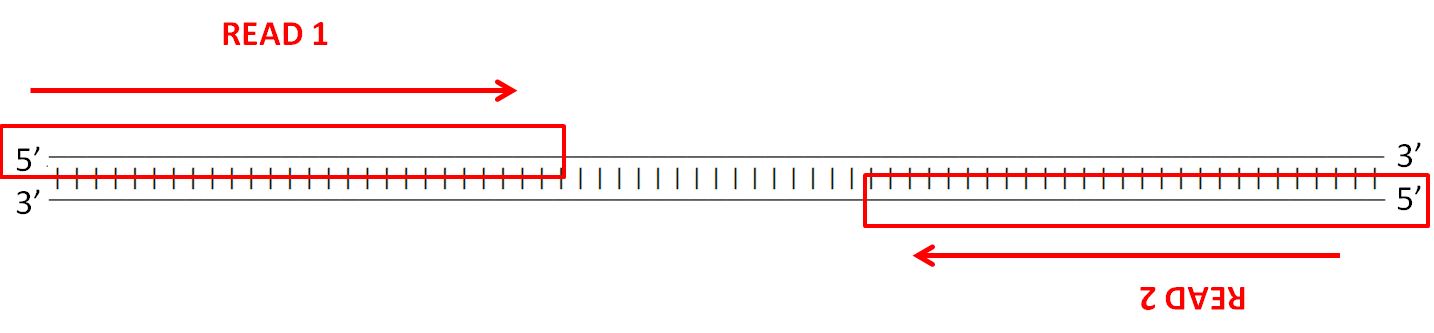
What 2 indicates in 2x150 bp, 2x300 bp maximum read length from Illumina website http://www.illumina.com/systems/sequencing-platforms.html?
I would be grateful if anybody can answer this?


I edited your post because you use (probably not desired) markup, e.g.
*will result in text in italics