Entering edit mode
7.2 years ago
mbk0asis
▴
680
Hi.
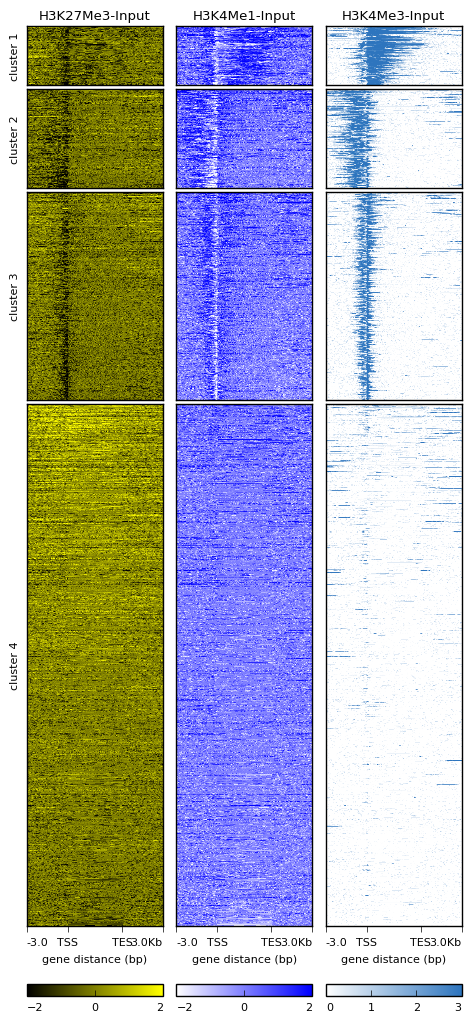
I generated a heatmap with several clusters using "deepTools" something like below, and I want to know the names of elements in each cluster. But, it seemed no tool existed among the "deepTools suit".
Can anyone help me?
Thank you!


Thank you! I missed that one...
BTW, if you have the matrix that produces the behavior you described here then feel free to send it to me.
I have the same question , I used this argument "--outFileSortedRegions" i get output something like this
I do have chromosome then the coordinates but apparently the names of the genes are missing why is it?
DeepTools cares about regions so it will typically ignore their names, which tend to be non-existent.
As Devon Ryan mentioned, gene names won't be appeared in the result. You can annotate the regions with gene names using 'bedtools intersect'.
I did annotate it with homer ,but i will check out the "bedtools intersect'" how to annotate it