We are happy to release PrimerServer, a high-throughput primer design and specificity-checking platform.
Project Page
https://github.com/billzt/PrimerServer
Demo Server
Pre-Print
Zhu T, Liang CZ, Meng ZG, Li YY, Wu YY, Guo SD and Zhang R (2017). PrimerServer: a high-throughput primer design and specificity-checking platform. bioRxiv 181941 (https://doi.org/10.1101/181941)
Description
Nowdays, it is easy to design one primer pair specific to the whole genome in a modal organism. Many programs such as NCBI primer-BLAST can conduct this well. However, what if you want to design multiple primer pairs that are specific to a custom genome? Besides, how to do this for both normal users (GUI needed) and bioinformatics users? (CLI needed, for large amount of sites). Here are several issues raised in BioStar:
- Primer Specificity
- Automated Primer-Blast like funcionnality
- PCR Primer Design - Specificity
- Pcr Primer In Highly Repetitive Region
- Design Pcr Primers To Uniquely Identify A Bacterial Strain
- Template-specific primers design
- ...
We anticipate that PrimerServer can satisfy all of this.
- Very easy to install. Only uses three tools: Primer3, Samtools and NCBI BLAST+. No other special modules or plugins needed.
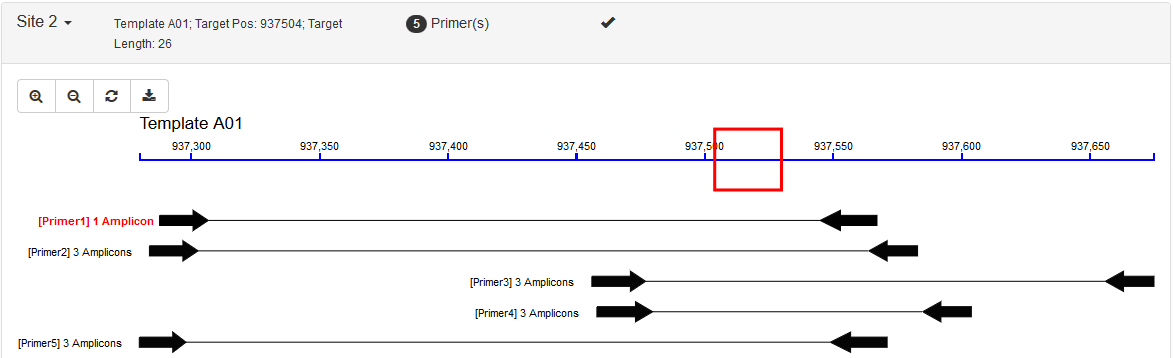
- Very nice GUI: Running progress bar, primer gragh, virtual gel electrophoresis, parameters and results storage
- Very fast CLI: multiple threaded, ~0.4s per site for designing primers for >10,000 sites
- High accuracy
For feedbacks, please use the GitHub tracking issues, or just send an email to zhutao@caas.cn
Thank you!

