Hello Biostars,
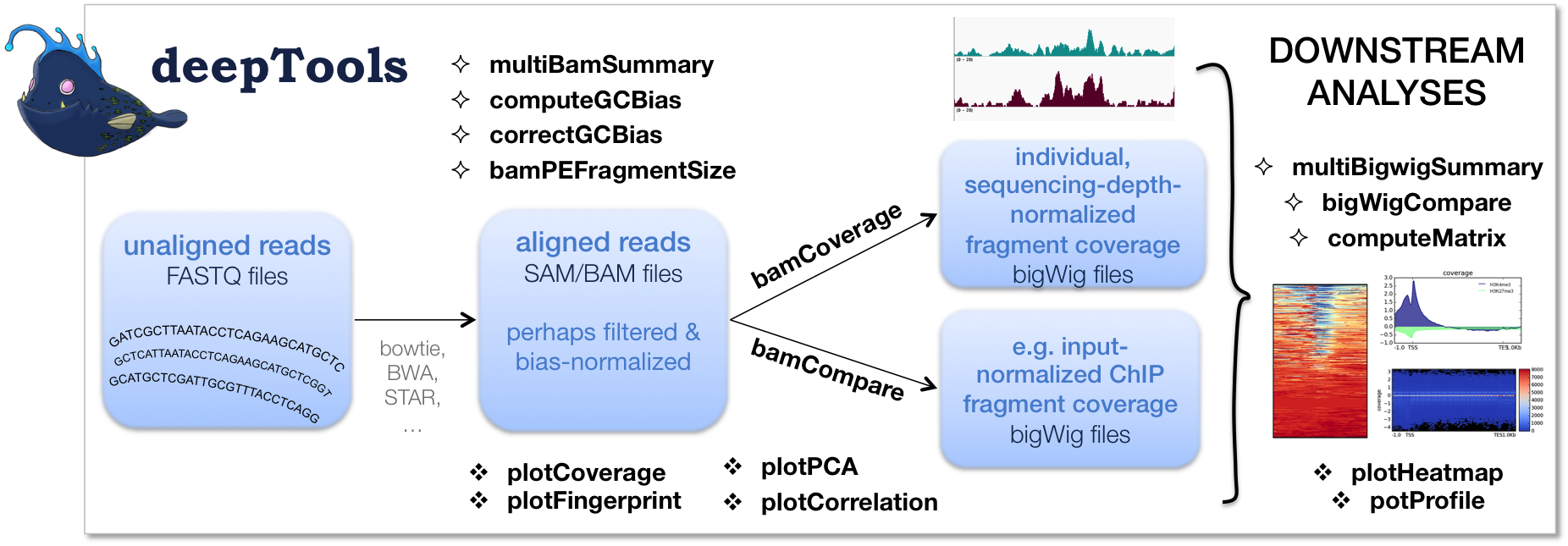
I need a graph showing the distribution of peaks near TSS and transcription termination site (TTS). Can anyone tell me any package/concept for it? I have tried ChIPseeker for TSS plot but I don't know how to plot TTS plot..
Please help me out. Thank you :)