I have a list of deferentially expressed genes and their gene counts obtained via RSEM/EBSeq. My raw data consists of RNA-seq reads from disease-free and relapse patients. When I make a heatmap, I can see that there are clusters of over or under expressed genes within each condition (relapse vs. disease-free). What tools exist to identify these clusters of genes? I believe Bioconductor's ConsensusClusterPlus is the only tool I'm familiar with. What other tools exist?
Note: I'm not seeking gene enrichment or over-representation tools, e.g. TopGo, ConcensusPathDB, DAVID, PANTHER. . . .

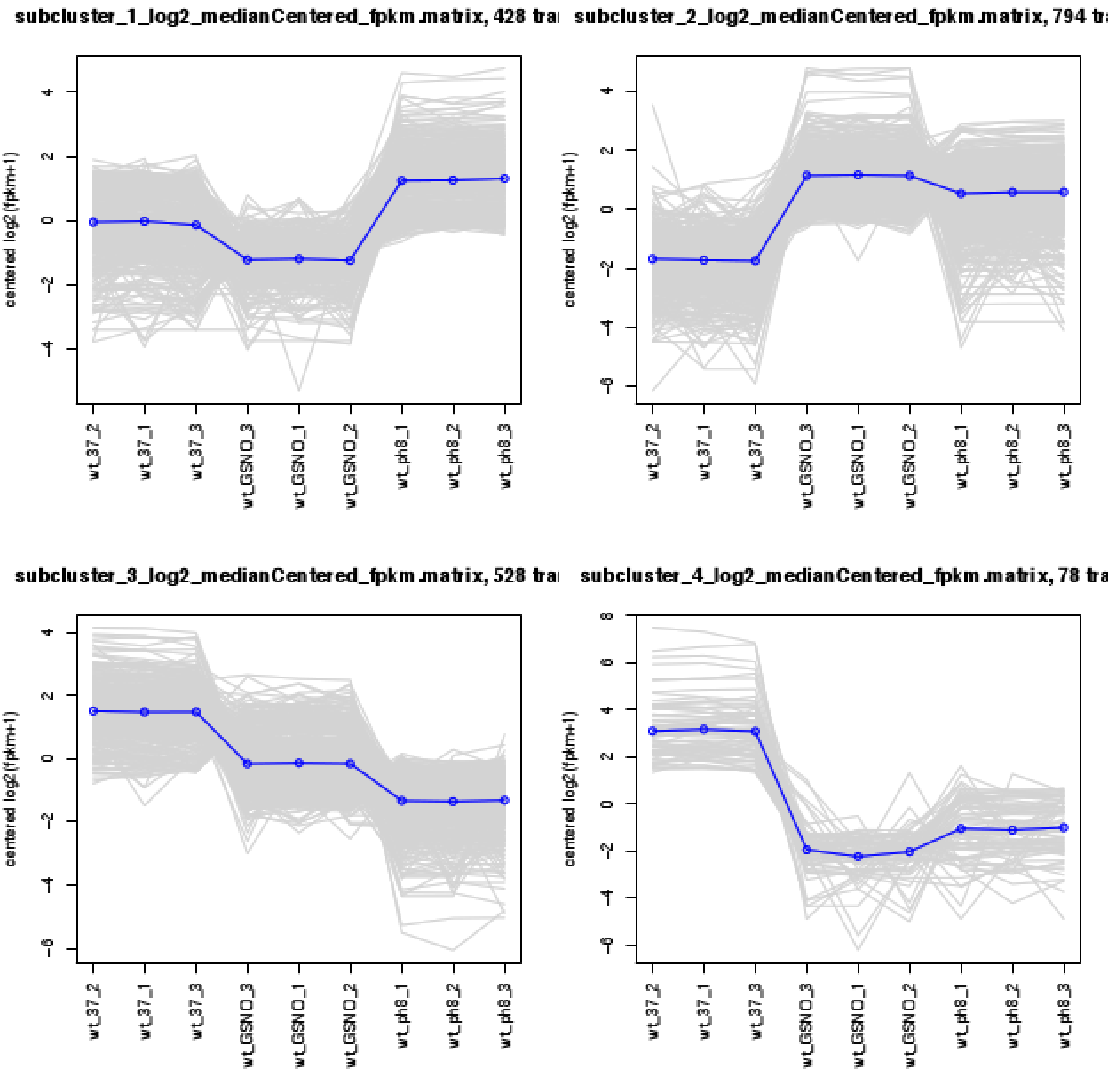

If you have large number of samples ( more than 15 samples per condition ) you can use WGCAN as mentioned by others to find modules of genes. But, if you dont have large samples, I would suggest to use simple clustering methods like hierarchical or k-means clustering. You can use different methods like elbow, or gap statistics to identify the possible number of clusters and use that information to create n-number of clusters by k-means.
Hi and thanks,
I have 3 replicates for condition A and 3 for condition B.