We just released the first public version of FlexiDot:
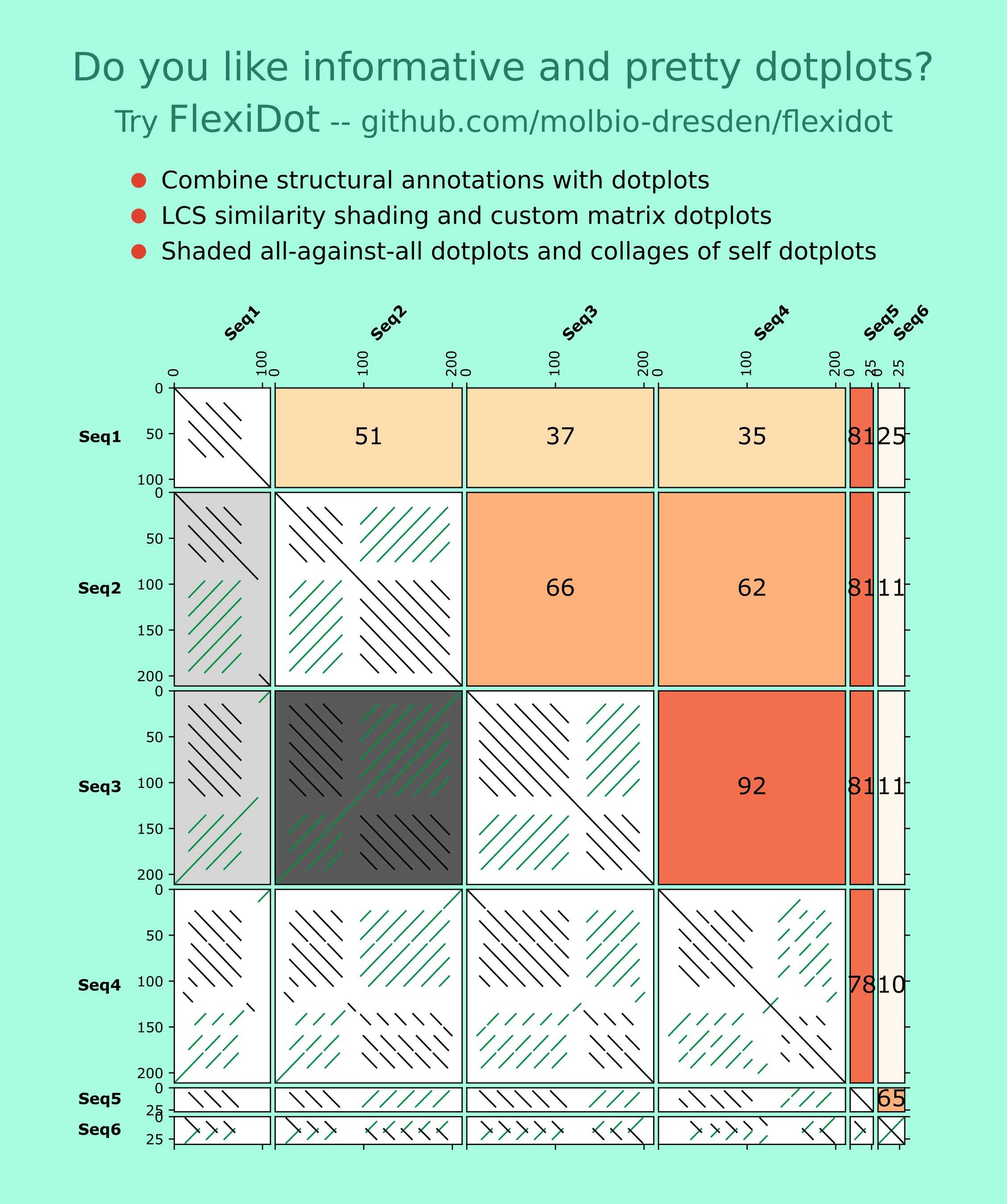
FlexiDot is a cross-platform dotplot suite generating high quality self, pairwise and all-against-all visualizations. To improve dotplot suitability for comparison of consensus and error-prone sequences, FlexiDot harbors routines for strict and relaxed handling of mismatches and ambiguous residues. The custom shading modules facilitate dotplot interpretation and motif identification by adding information on sequence annotations and sequence similarities to the images. Combined with collage-like outputs, FlexiDot supports simultaneous visual screening of a large sequence sets, allowing dotplot use for routine screening.
Some FlexiDot features:
- handling of error-prone SMRT reads and ambiguity-containing consensus sequences (e.g. derived by alignment or assembly).
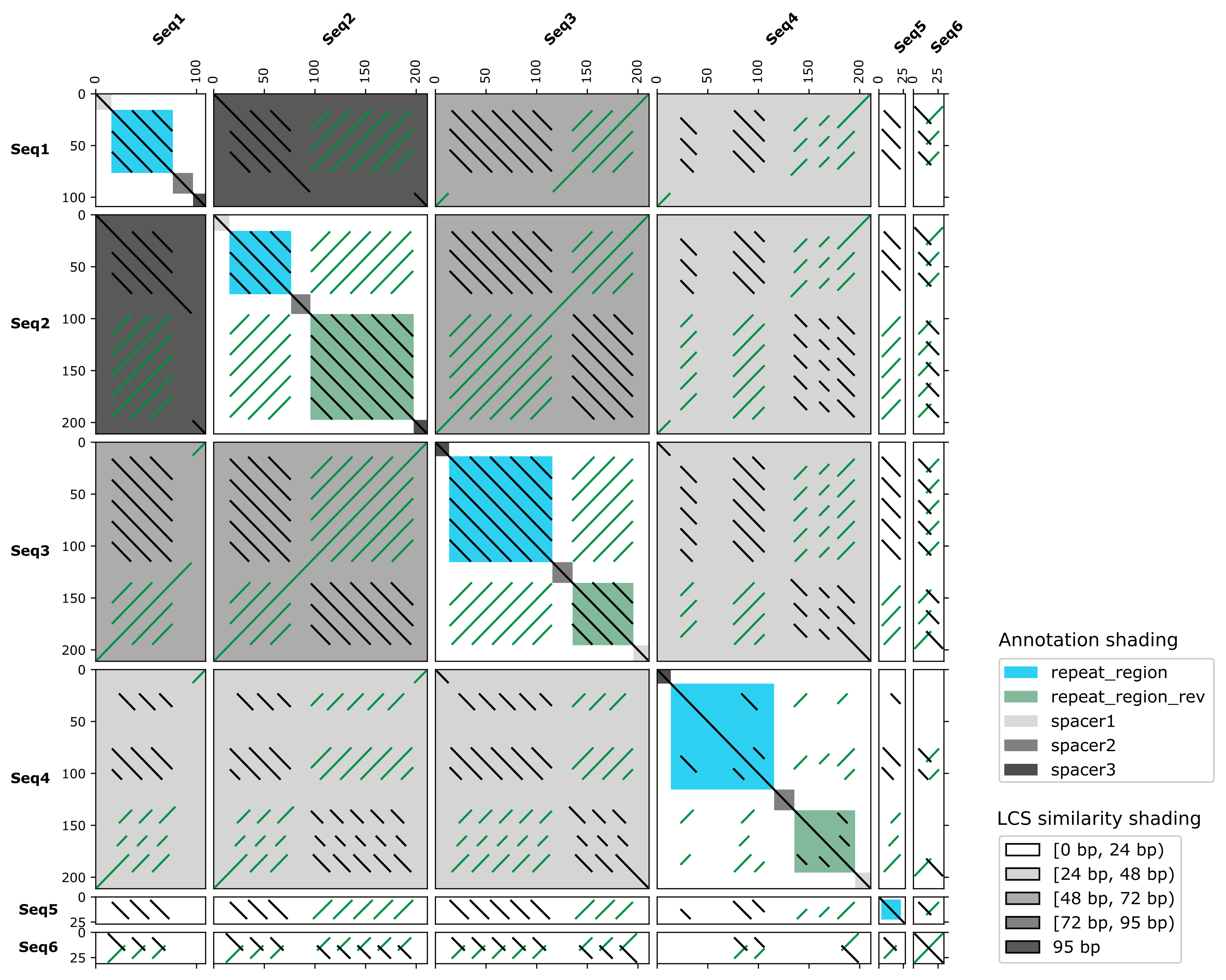
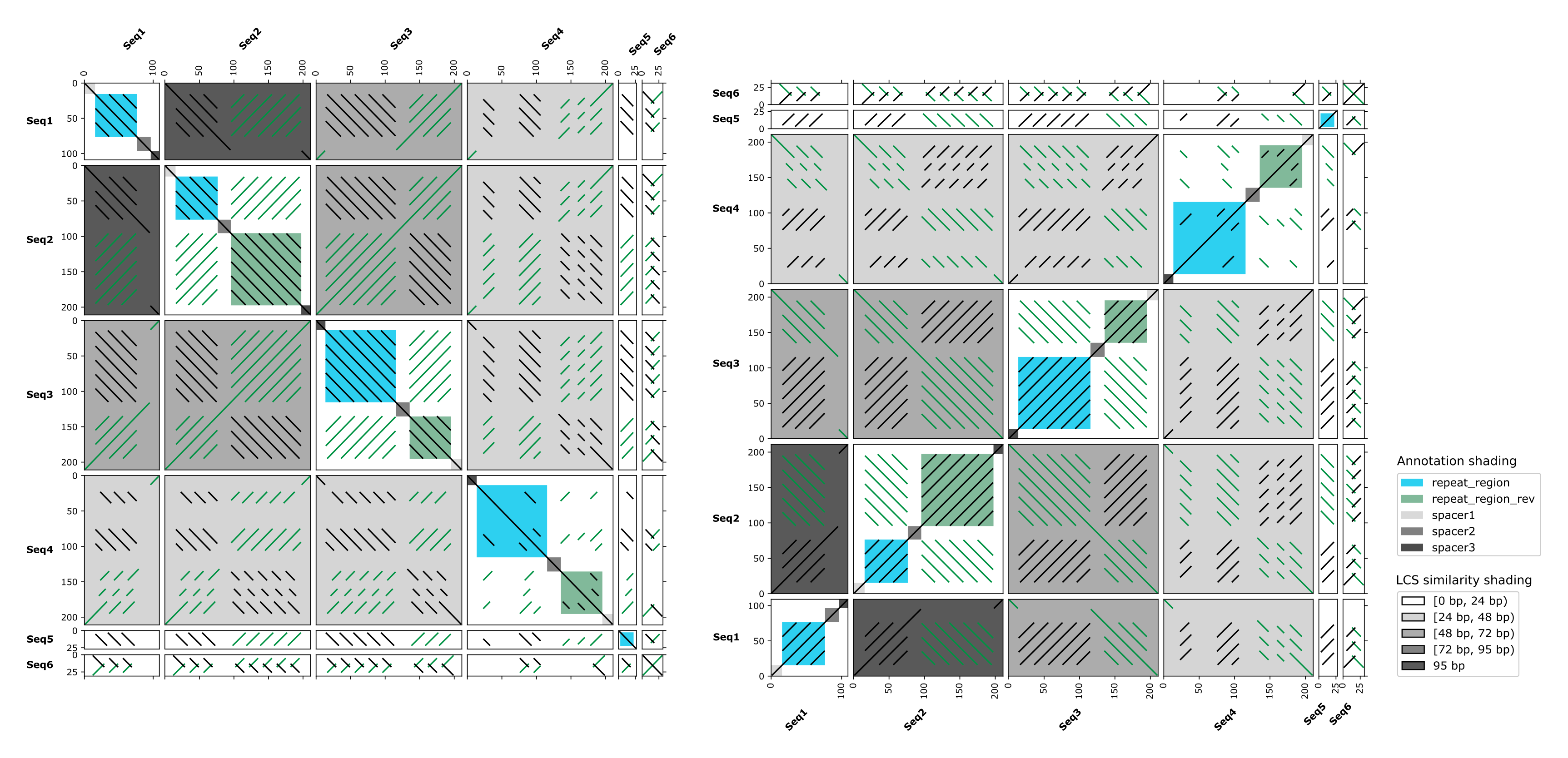
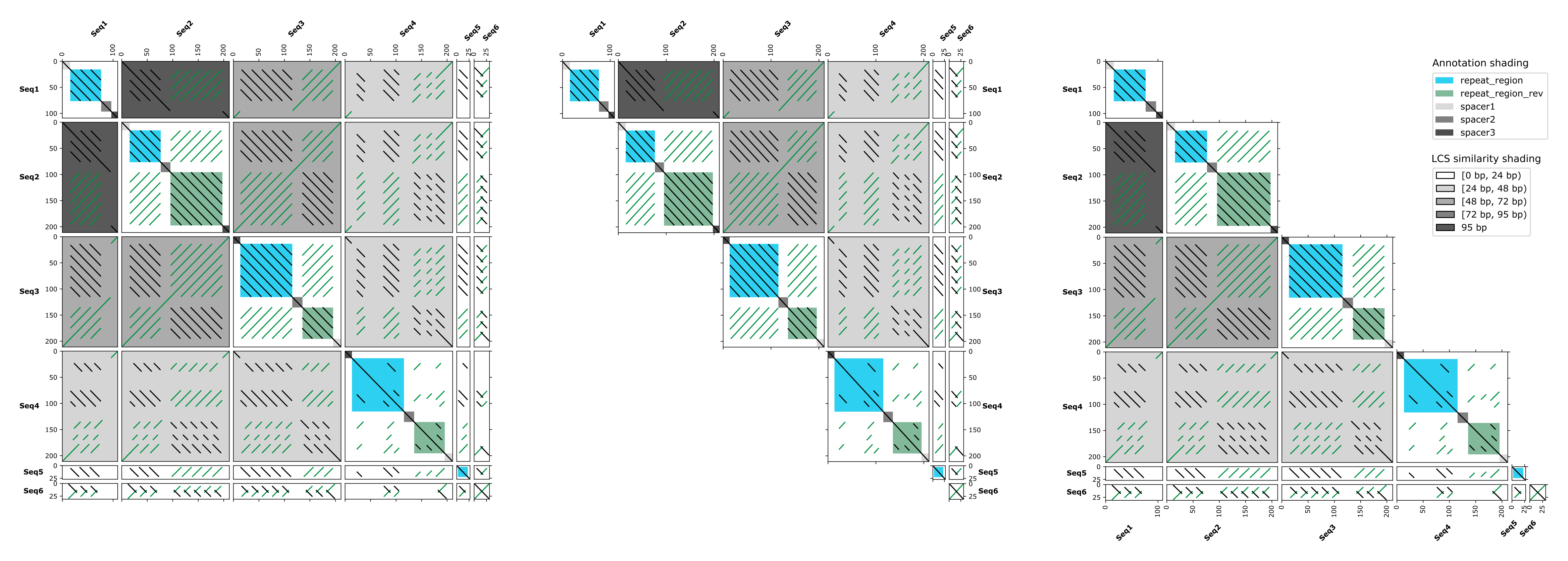
- integration of descriptive information on the analyzed sequences (e.g. gff3-type structural sequence annotation or pairwise identities)
- similarity shading modes
- high flexibility for customization and automation
- output as vector and raster graphics
- self, pairwise and all-against-all visualizations
See for example this tutorial on adding gff3-type annotation to a dotplot.
More:
Code, examples and documentation on github: http://github.com/molbio-dresden/flexidot
Publication: K. M. Seibt, T. Schmidt, and T. Heitkam (2018) "FlexiDot: Highly customizable, ambiguity-aware dotplots for visual sequence analyses". Bioinformatics, in press, http://doi.org/10.1093/bioinformatics/bty395
(Preprint: Latex version at Overleaf)


IMO, the best software for creating dotplots.
Thank you a lot for saying this. It makes us happy, since we also love this tool.
It would be great to have an option to display dotplots according to cartesian coordinate system (i.e., Y axis from bottom to top, rather than from top to bottom).
Thank you for the suggestion! Some tools as e.g. the EMBOSS polydot package handle plots just like you suggested. (And we even actually plotted like this in very early FlexiDot versions, but then changed this, because most use it the other way round.)
I spoke to Kathrin (who is first authoring FlexiDot), if we want to do implement this - and she told me that it is already done! She just couldn't resist. So... we will have to rigorously test that everything behaves well, and then you'll probably get your very own -M (mirror) parameter. ;)
Wow! That's great news. I'm looking forward to it. My best wishes to the entire FlexiDot team.
Hello a.zielezinski, we just put forward FlexiDot v1.04 containing your suggested parameter
-M/--mirror!Thank you very much. This feature is very helpful. I'm already showing FlexiDot to my students.