Hello,
I have trimmed my sequences with TRIMMOMATIC using the following command:
trimmomatic.sh PE -phred33 -threads 8 <seq_1.fq.gz> <seq_2.fq.gz> <seq_1_paired.fq.gz> <seq_1_unpaired.fq.gz> <seq_2_paired.fq.gz> <seq_2_unpaired.fq.gz> LEADING:13 TRAILING:13 SLIDINGWINDOW:7:30 MINLEN:30
I think the trimming has been a good thing to do because the quality control (performed with FASTQC) shows slightly better sequences, with higher quality overall and lower warning flags. For instance, the original seq_1 has the following features:

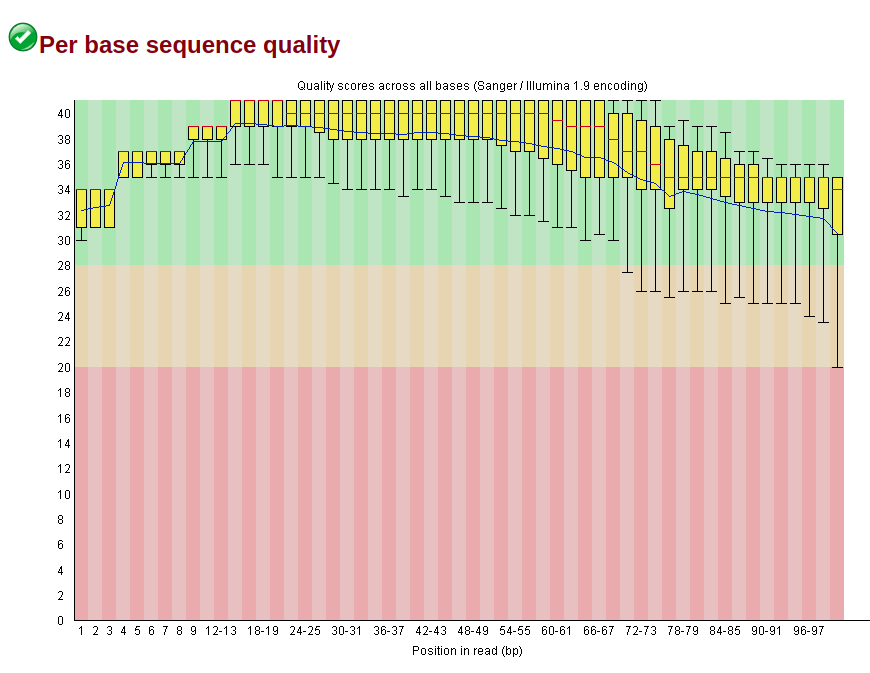
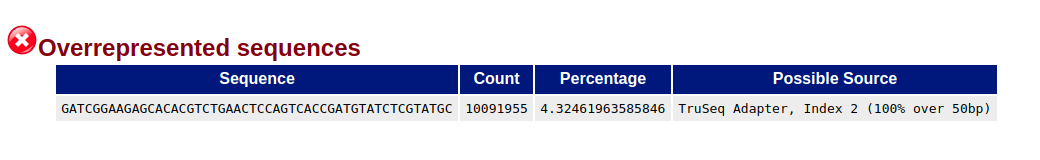
 The file also shows the presence of the adaptors, even if they do not show in the adapter content flag (maybe because Trueseq is not listed in the graph):
The file also shows the presence of the adaptors, even if they do not show in the adapter content flag (maybe because Trueseq is not listed in the graph):
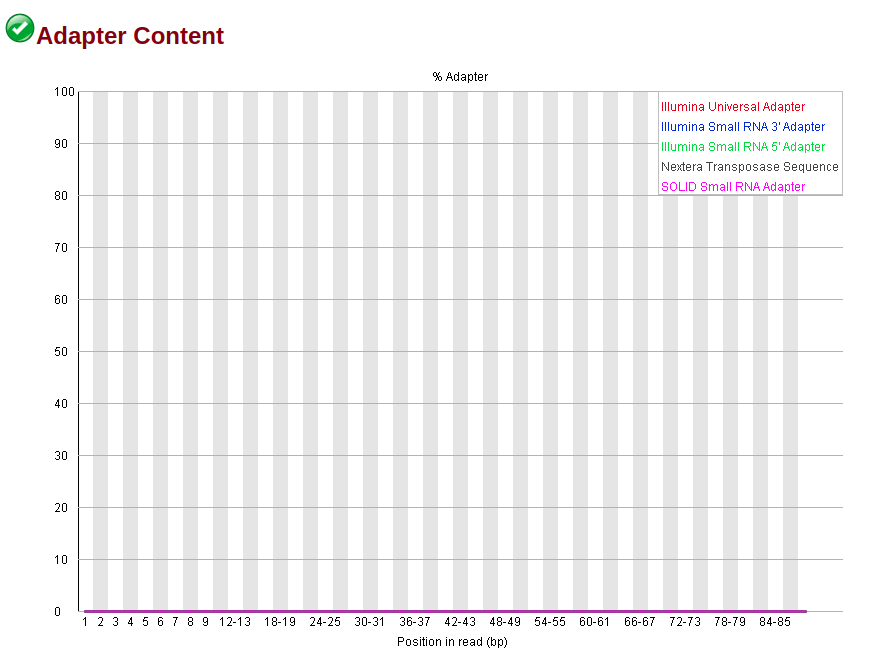
The trimmed sequence gives:
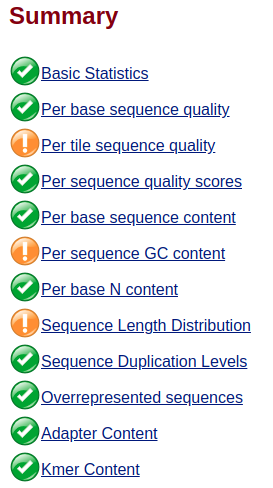
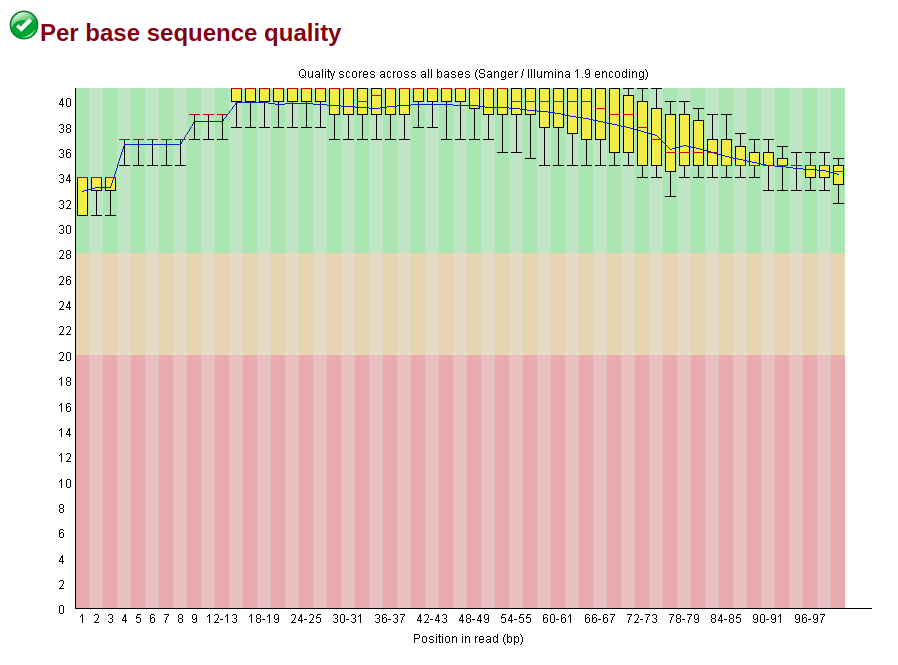
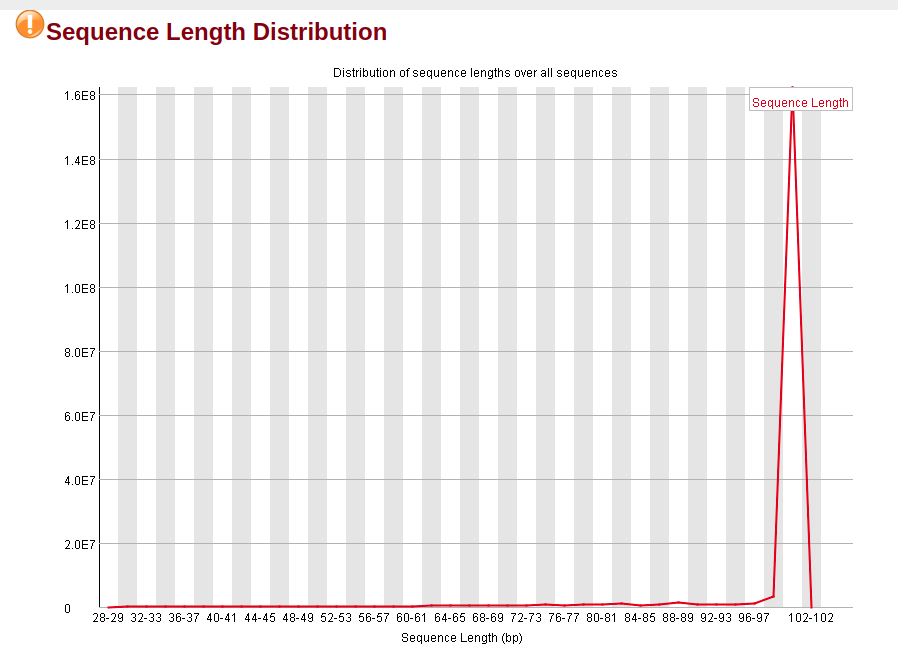
To note that both sequences have a GC and per tile warnings (nothing much I can do there, is it?), but the trimmed does not have the adapter and k-mer warnings anymore, along with higher per base quality.
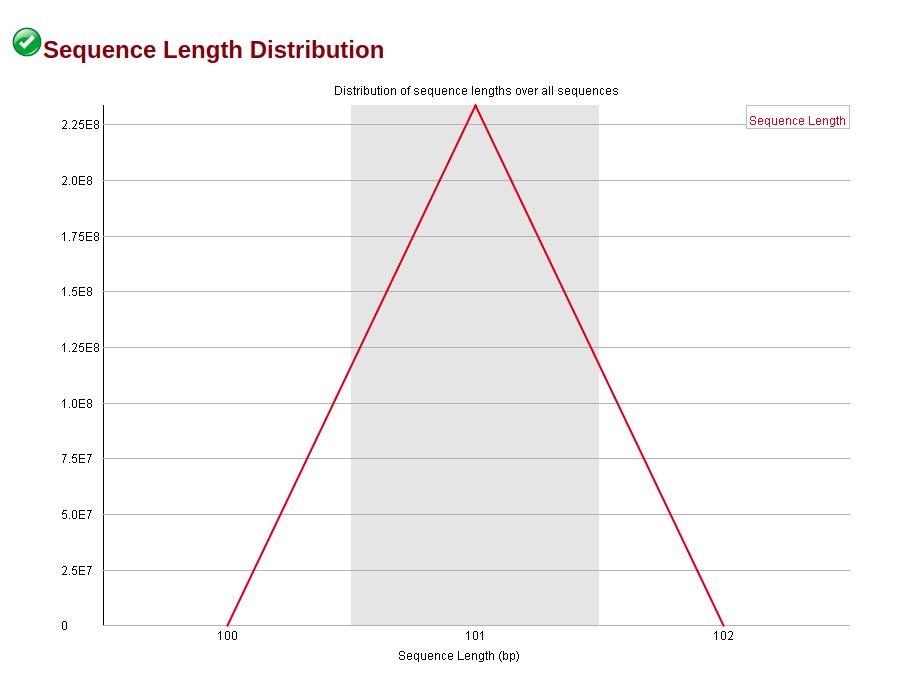
HOWEVER, the trimmed sequence has the Sequence Length Distribution flagged. Can I simply ignore this flag, since I think is coming out simply from the fact that I reduced the reads' length?
Thank you.


I've moved this to an answer, since it's absolutely correct.