Hello
I was wondering whether there is an Illumina WGS dataset (fatsq files) that is quality control flaw-less, something that does not have yellow of red flags in any of the FASTQC fields, no tile errors or kmers, for instance but shows the typical decreased of quality values at the extremities. And if yes, does it have a SRA number or a website where can I download it?
Thank you


What are you actually looking for? What is your objective with this dataset, if it exists?
I suppose you could take any reasonably good dataset and apply some filters to get that "perfect" dataset.
That said, the colours in fastqc are just indications and do not determine conclusively if a dataset is appropriate for a biological question.
I just need a didactical dataset that shows how fastqc works, but all the datasets I have available show some defect of some sort. I still haven't found one that does not have any flag raised. I need to produce a figure like that one sees in the manual, but without a good dataset, I cannot. I could trim, but in that case, it would not be an original set...
I even got a dataset reported from a manual (I won't say which one) and the base quality figure is as good as in the book; what they did not show was the associated summary: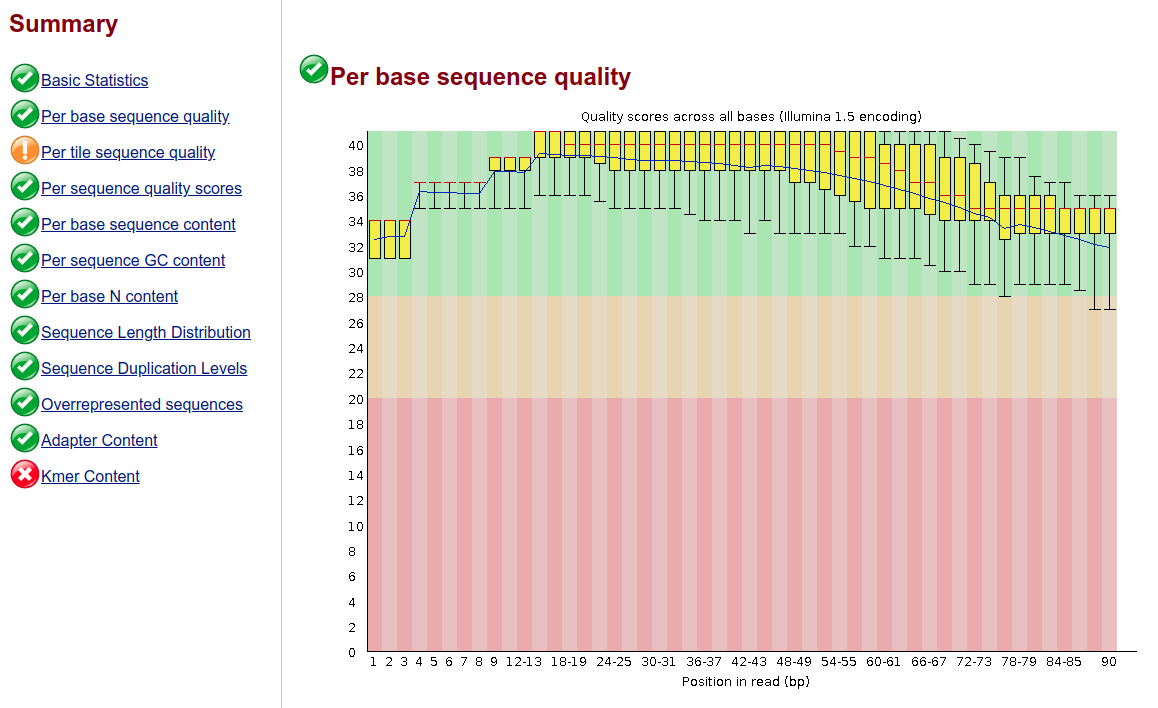
also, this was an exome analysis, not WGS...
I'm not even sure what would be required to have every single fastqc flag pass. I've never seen a green tick for Kmer content.
FastQC has a file called
limits.txtthat you will find in theconfigurationfolder in FastQC distribution. If the redX'sbother you that much feel free to edit and change the intervals in this file (that throw those red X warnings) so everything becomes green.As others have said there are no perfect datasets. It is important to keep the context of the experiment in mind as you look at FastQC results. Use the results as a guide to decide if you should do anything additional to the data (e.g. trim, normalize etc) or just proceed with your usual analysis workflow. You will know a really bad dataset (that you should discard) when you find it.