I have a patient with a homozygous insertion, although I can detect the presence of the insertion using the software that I have, it is not so clear as to whether or not the individual is homozygous for this insertion. Is there any way this can be detected using sanger data?
Yes.
- The Sanger sequencing trace will look very 'clean', with only single allele peaks on the chromatogram at each nucleotide (see the first 8 nucleotides of ATpoint's figure above), but there will be some additional bases compared to the reference at the site of the insertion.
Some other points worth noting:
Increase the size of your PCR amplicon to include known common polymorphisms, and if you are lucky, one of them may show as heterozygous on your Sanger sequencing, confirming both alleles have amplified.
Be careful that your PCR or sequencing primer does not anneal over a common polymorphism. This can result in you inadvertently amplifying only one allele - the one with the insertion - and it will look homozygous. This is called allele drop out, I think.
If the insertion is rare, sequence the parents, as both may be heterozygous. (Although one or both parent may be homozygous if it is a common insertion).
A heterozygous insertion will look like this;
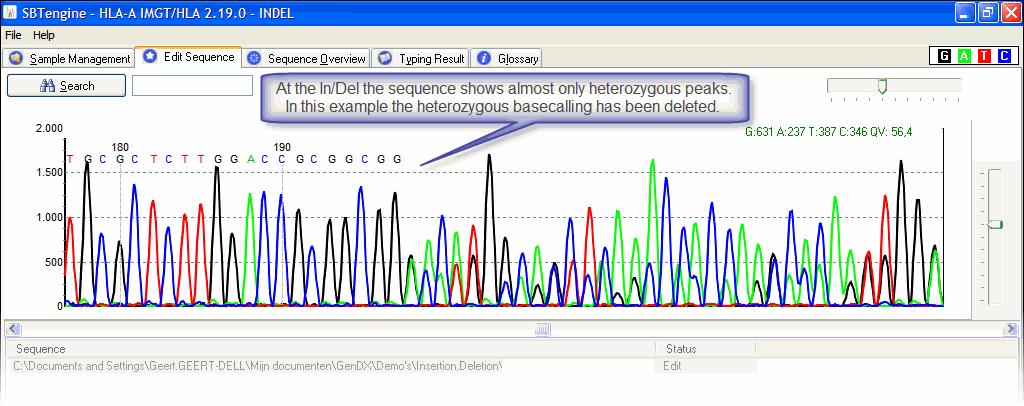
A homozygous one will look like a regular electropherogram.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


I would look at the raw chromatogram and zoom to the insertion site. If it is heterozygous, you should see two signals coming up. Programs like SnapGeneViewer can visualize the
.ab1files. Heterozygosity would look like this:Correct. But this is a homozygous insertion and there would only be one peak at this site. I need to confirm that both alleles have the insertion
Well, if you see the additional nucleotides compared to the reference, and clean single peaks, then it should be homozygous.