Entering edit mode
5.7 years ago
roy.granit
▴
880
Hi All,
I have matrix with SNP calling for several positions in 6 bacterial genomes relative to a ref. sample. (generated using snippy)
For example:
gene position wt sample_1 sample_2 ...
nm_1234 123 A C T
nm_1234 129 G T A
Now my goal is to find the distance between the different species and display it on a nice plot like this :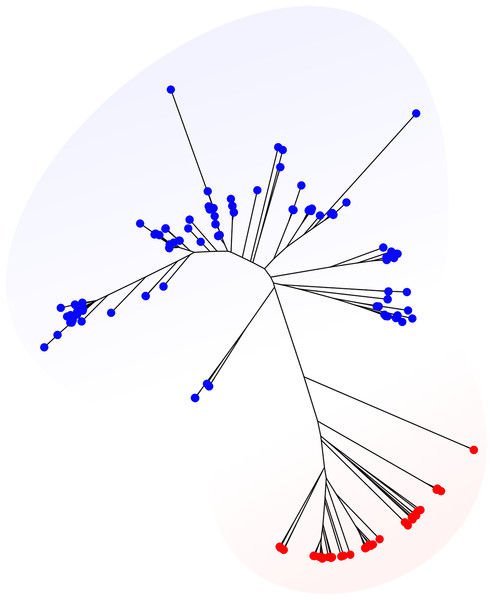
Any suggestions on hoe to approach this? Thanks!


Past threads that may be useful:
Thanks. I was able to use FastTree to convert the data into Newick format, now I need to find a way to plot it was nicely as in the figure.. would be happy to receive suggestions
After creating Newick file ,I used Dendroscope to create the "radial" plot