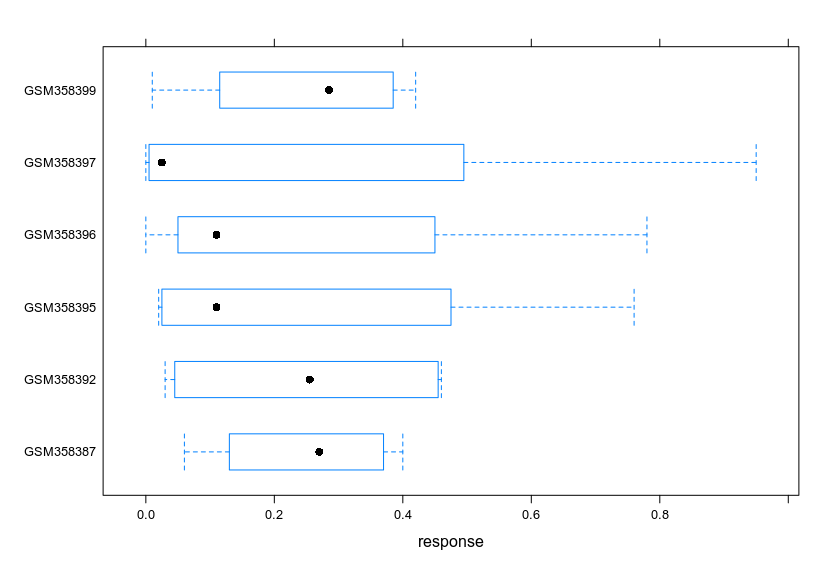
How do I boxplot each row in a dataset in R?
In the dataset below, I want to plot RF.CMS1.posteriorProb, RF.CMS2.posteriorProb, RF.CMS3.posteriorProb and RF.CMS4.posteriorProb for each GSM sample in column X. So separate boxplots for each row in column X, in R.
Appreciate any help in this regard.Many thanks.
head(GSE14333_pheno_new)
X Location DukesStage Age Gender DFSTime DFS_group DFSCens AdjXRT AdjCTX
1 GSM358387 Rectum B 54 M 9.96 poor 0 Y Y
2 GSM358392 Right B 38 F 17.95 poor 1 N Y
3 GSM358395 Right B 78 F 22.02 poor 1 N Y
4 GSM358396 Left B 65 F 22.38 poor 0 Y Y
5 GSM358397 Left B 65 F 22.38 poor 0 Y Y
6 GSM358399 Left B 56 F 25.21 poor 0 Y Y
RF.CMS1.posteriorProb RF.CMS2.posteriorProb RF.CMS3.posteriorProb RF.CMS4.posteriorProb
1 0.20 0.34 0.40 0.06
2 0.46 0.06 0.03 0.45
3 0.76 0.02 0.03 0.19
4 0.10 0.78 0.00 0.12
5 0.01 0.95 0.04 0.00
6 0.35 0.42 0.22 0.01
RF.nearestCMS RF.predictedCMS predict.label2 dist.to.template dist.to.cls1.rank nominal.p
1 CMS3 <NA> CRIS-B 0.7331209 68 0.00019996
2 CMS1 <NA> CRIS-A 0.8965833 52 0.00739852
3 CMS1 CMS1 CRIS-B 0.8559375 80 0.00019996
4 CMS2 CMS2 CRIS-C 0.7944693 111 0.00019996
5 CMS2 CMS2 CRIS-C 0.8465627 120 0.00179964
6 CMS2 <NA> CRIS-D 0.9366855 148 0.00719856
BH.FDR Bonferroni.p
1 0.0006725928 0.0369926
2 0.0102143750 1.0000000
3 0.0006725928 0.0369926
4 0.0006725928 0.0369926
5 0.0026849469 0.3329334
6 0.0100130350 1.0000000


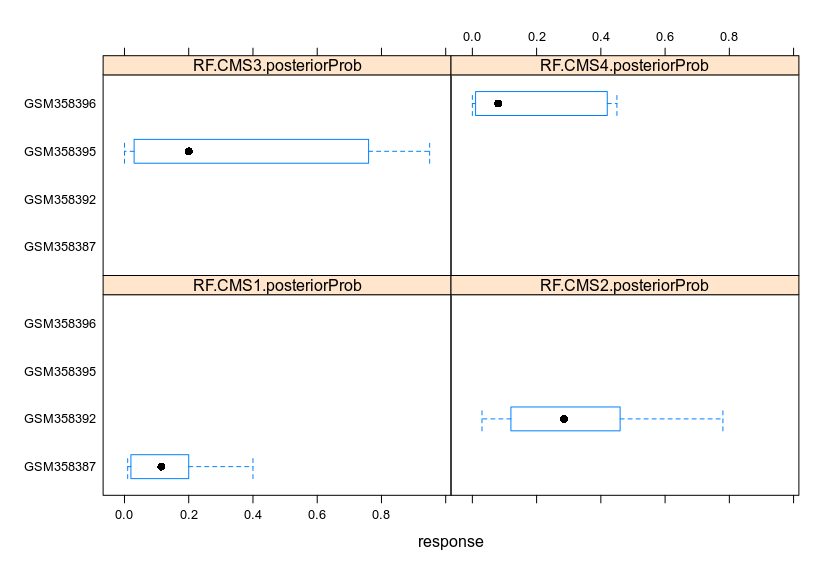 Lattice facilitates the clustering of data. Changing the parameters allows to cluster the data to fit the demand.
Lattice facilitates the clustering of data. Changing the parameters allows to cluster the data to fit the demand.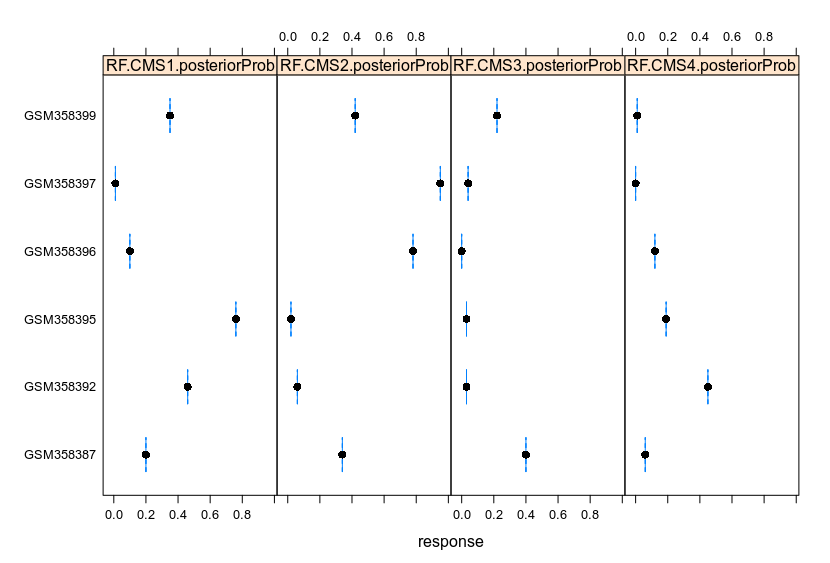

bio94 : If an answer was helpful, you should upvote it; if the answer resolved your question, you should mark it as accepted. You can accept more than one if they work.
