I am running MaSuRCA 3.2.3 for genome assembly but getting some error as follows.
[Sat Nov 3 15:05:16 IST 2018] Processing pe library reads [Sat Nov 3 15:05:17 IST 2018] Average PE read length 150 [Sat Nov 3 15:05:18 IST 2018] Using kmer size of 105 for the graph MIN_Q_CHAR: 33 Estimated genome size: 340625642 [Sat Nov 3 15:05:18 IST 2018] Computing super reads from PE [Sat Nov 3 15:05:18 IST 2018] Super reads failed, check super1.err and files in ./work1/
Super reads failed, check super1.err and files in ./work1/ following the instruction i checked super1.err file
super1.err file contains :-
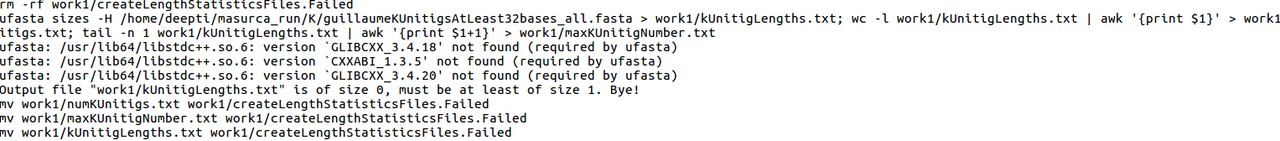 my commands are as follows :-
my commands are as follows :-
masurca config -o assemble.sh -l /usr/lib64:/usr/lib
./assemble.sh
and the config file and assemble file is attached here.
Can you please help me out from this issue as soon as possible. thanks with regards


Have you tried recompiling (rerunning install.sh)? Looks like ufasta is looking for GLIBC libraries that aren't there