Entering edit mode
8.7 years ago
mangfu100
▴
800
Hi all.
I have some questions regarding the distribution of copy number variations.
While reading some papers, I found that they often grouped the CNVs according to its size.
As shown in figure a, this paper used a normalized chromosome arms to separate focal and arm-level SCNVs.
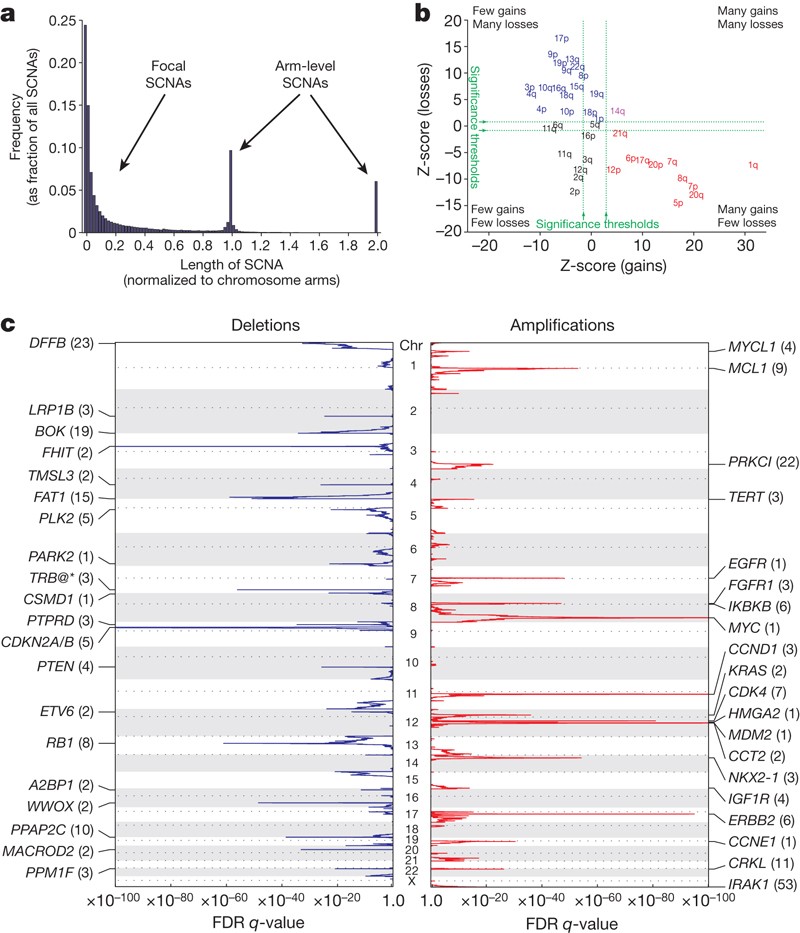
However, I don't know how to normalize chromosome arms. Does this mean the Z-score of each segment's length? I don't think so.
Can you help me to measure the normalize CNVs according to chromosome arms in detail?


I also have this question