Hallo we have an RNA-Seq experiment and I would like to ask for your suggestion about how to best sequence the samples
We are working on mammalian cell lines (human or mouse) and would like to sequence samples for a time-series analysis
we have four conditions and four time-points each in triplicates. In total we have 48 samples. We have a nextSeq 500 and we'll use the high kit with ~400M reads.
As we would like to reach a good coverage we decided to split the samples on two (or maybe three cell lines). If we take three flow-cells there won't be a problem, as I can just put one replicate of each of the three on one flow-cell.
My question is what would be best to do, if using only two flow-cells? we would than like to put 24 samples per flow cell. I was thinking to run it like this-
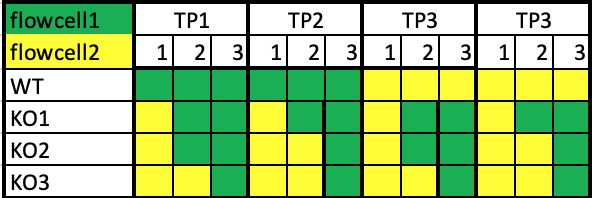
Would it make sense, or do I miss some important factors?
thanks


Can you have 48 barcodes in one flow cell run on a nextseq 500?
Yes, there is no limitation to my knowledge as long as you have primers with enough barcode combinations. A dual-indexing would be a good choice here. Pooling everything is also actually the "most correct" way of dealing with this problem, as any flow cell-specific confounding effect would equally affect all samples.
This is exactly my question. Can I use the nextseq 500 with enough unique primers/barcodes for 48 samples? which kit would it than be?
I double-checked with the other moderators and they confirm that there is no technical limitation towards the number of barcodes for Illumina machines. When you perform your indexing PCRs, you could use the Dual-Indexing Multiplex Oligos from NEB (or any other company), which allows combinations for up to 96 samples and is compatible to standard TruSeq library prep. I hope i got your "which kit" question right.
yes thanks. I'll try it.
@ genomax Is this same as combining two runs to increase the coverage? One case I encountered is as follows:
Core argument is that there are enough examples on pooling lanes, runs to remove batch effect. This way they can merge data between runs.
If the same pool is being sequenced multiple times there should be little or no batch effect (unless you are switching sequencer chemistry/type).