Hello I am sort of a newbie in statistical science, so I hope this isn't a terrible question.
I am doing an analysis on example data, and I do the MAplot on my normalized counts from my DESeq2 results. This is what I get:

So we have a normal(i think) MAplot, of my two groups, each group has 3 samples.
But in bottom left corner, that is downregulated genes with low read counts we have a linear relation between read counts and Logfold change.
What could cause this?
Are these genes valid?
Another similar phenomena is seen when you compare two samples counts(before any rlog transform):
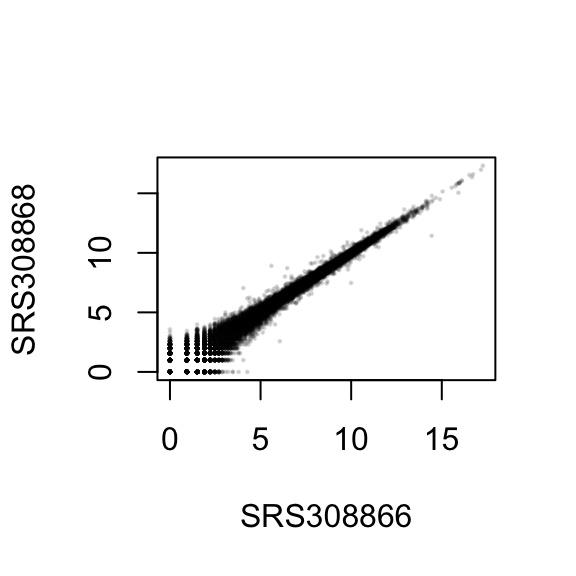
At the low read counts, again we have what looks like straight lines. Does it have something to do with the low sampling rate(counts) of these genes causing aliasing?
Best regards, Gabriel


How to add images to a Biostars post