I have aligned the small RNA reads with reference genome, now I want to know the method to count the total number of small RNA both (novel and know) in my reads? any method or tool please mention here, I will be thankful.
Hi,
You can have a look at Genboree's exRNA Tools. They offer it as a service, but also disclose what they are doing (see below). Nevertheless, novel miRNA detection needs a bit more analysis, e.g. as in mirTools 2.0
Cheers,
Michael
It was first developed for plant genomes, but I've had very good experience with ShortStack from the Axtell lab with both Arabidopsis and human smRNA-seq. They use weighting to assign multimappers (a vast majority of reads when performing small RNA-seq) to obtain more accurate counts for small RNA "clusters." You can then intersect these clusters with miRBase to see which correspond to annotated miRNAs and which are potentially novel.
A schematic of their mapping algorithm is below, and the publication is available here.

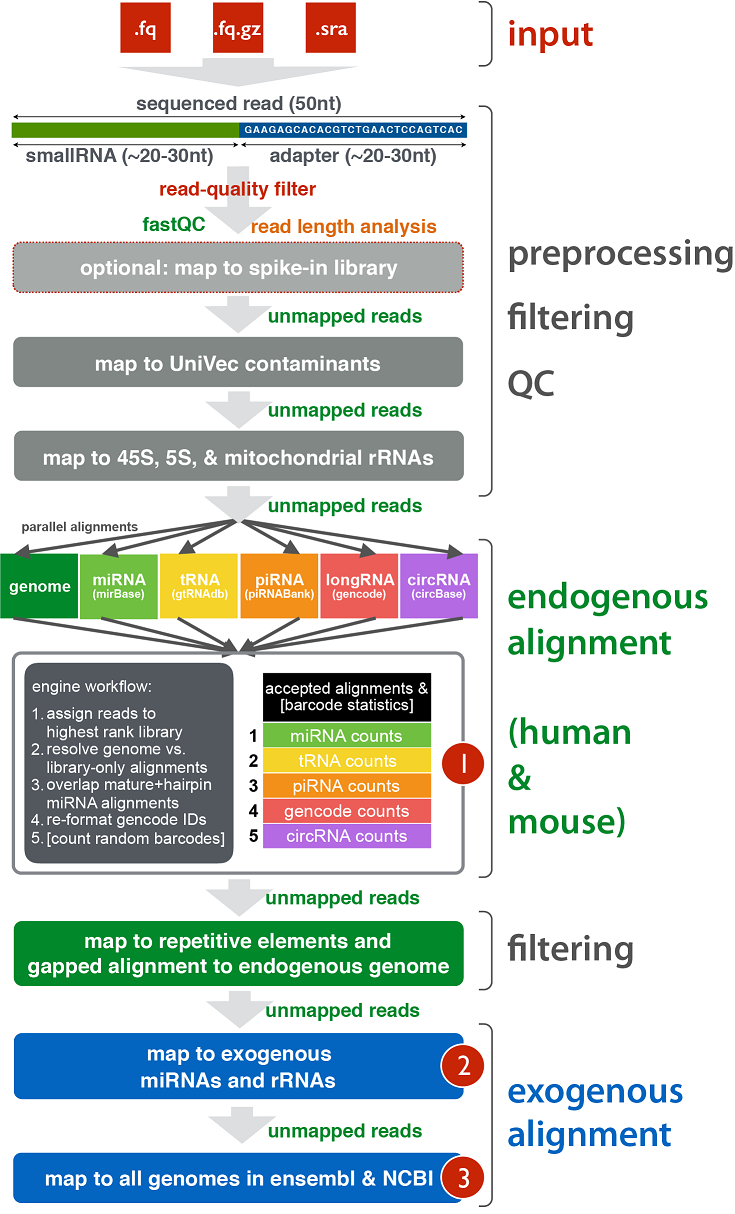
Hi,
If you do not care about the aligner you are using, we have recently implemented an algorithm that hopefully improves the quantification of small RNA reads (and uses bowtie1 as a default aligner). The tool is called Manatee and the related paper is available here.
Cheers,
Joanna
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Depending on your genome you can use an applicable GFF file available at RNAcentral to count aligned reads using
featureCounts.