Hi,
I would like to use Harmony to remove batch effects from my 10x Genomics scRNA-seq data combined from three donors. However, I am wondering whether Harmony requires the same cell populations to be present in all samples, or how it deals with a population that is e.g. unique to one donor?
Many thanks,
Lucy

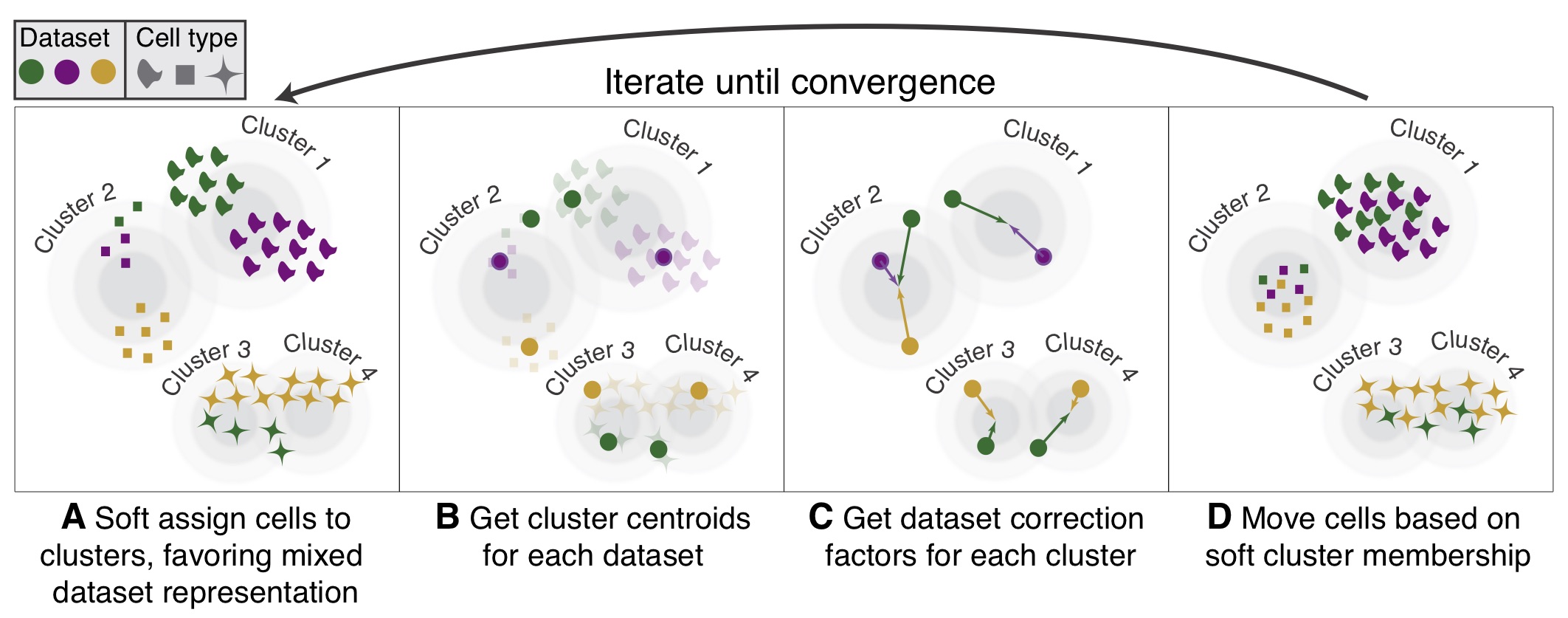

looks like it requires same population.