For my own ancestry results, I would say that the broad-level assignments (like the 1000 Genomes super-populations) were accurate but I had some concerns about some more specific predictions and smaller segments.
I have already gotten some useful feedback from the community through various means (even though some of those are for other topics). However, I wanted to make sure I was providing a fair mix of potential utility as well as limitations.
I expect that there are probably some general ideas about ancestry / race / ethnicity that may be important to communicate to the general public. For example, I think some Twitter threads from others match my feelings (such as this and this). However, I also thought that perhaps it might help if this discussion could relate to some more specific examples.
So, in terms of some specifics for my own results:
- On the positive side, I found slightly more distant relatives (second cousin or closer) whose relationship I could confirm by talking to other relatives (although I would also say “relatedness” is a little different than “ancestry”). However, I feel differently about those that are closer than a 4th cousin versus more distant cousins.
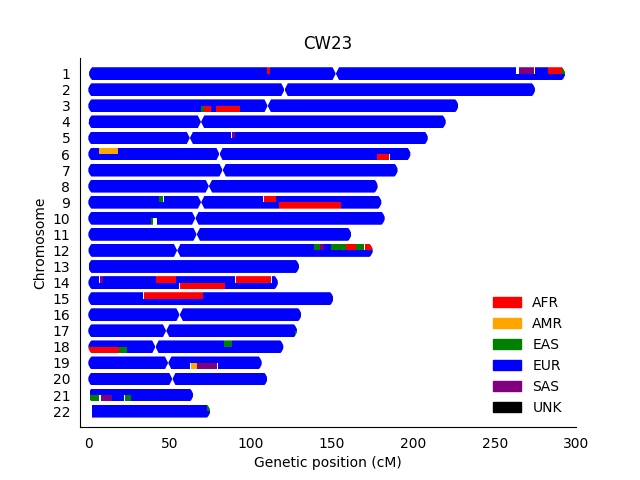
- I have approximately 2% African ancestry predicted from my father’s side. So, I think there should probably be a larger segment on one copy of my chr14. However, with RFMix, the African (AFR) ancestry was split between copies in nearby locations, which I would guess was due to phasing issues:
- Also, as mentioned in the first blog post, I noticed that I could filter some likely false positive results on 23andMe if I increased the confidence threshold from 50% to 90%. Do you agree with the troubleshooting strategies that I discuss in that post, or do you have other suggestions about what somebody can do to critically assess a surprising result?
- For example, have you found it useful to re-analyze raw results with tools like GEDmatch, impute.me, DNA.land, GENOtation, MySeq, etc.?
While I would like to keep discussion as public as possible, I did encounter at least one individual that shared at least one significant segment (identified through GEDmatch) where I didn’t really want to give out the information that might help interpret that signal. So, it is possible that I might wish to switch some conversations to more private communication (like e-mail or a Twitter Direct Message), but I will try to maximize what I answer publicly.
Do others have examples of either successful confirmation or false positives in their own results?
Thank you in advance for your feedback!

