Entering edit mode
4.1 years ago
rioualen
▴
710
Hi, I'm looking for programs to generate heatmaps from DNA sequences (TF binding sites) and clusterize them. I like this type of figure but it's from a Matlab library, and I haven't found equivalents in R or else. Any suggestions? Thanks a lot!
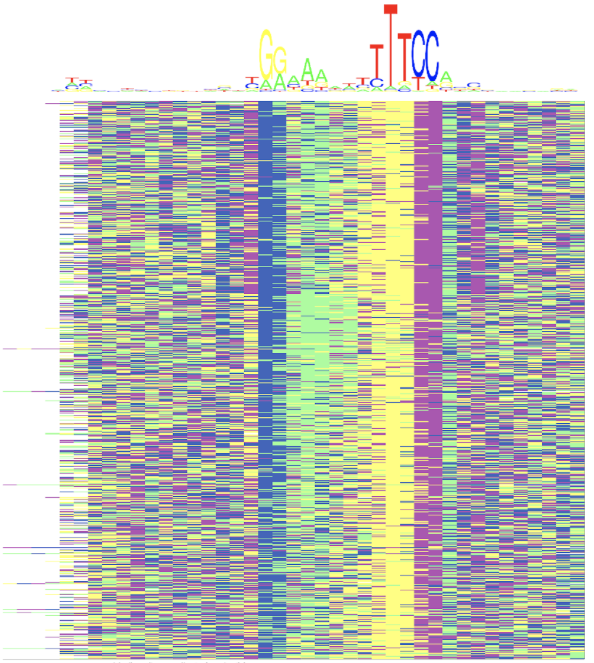


Yeah I know it, but I'm not looking to produce the logo, I'm rather interested in how the sequences possibly cluster into subgroups, or if they're more like a continuum.
So you can represent the MSA as a matrix and you plain heatmap