Dear all,
I am a beginner, sorry if too simple question.
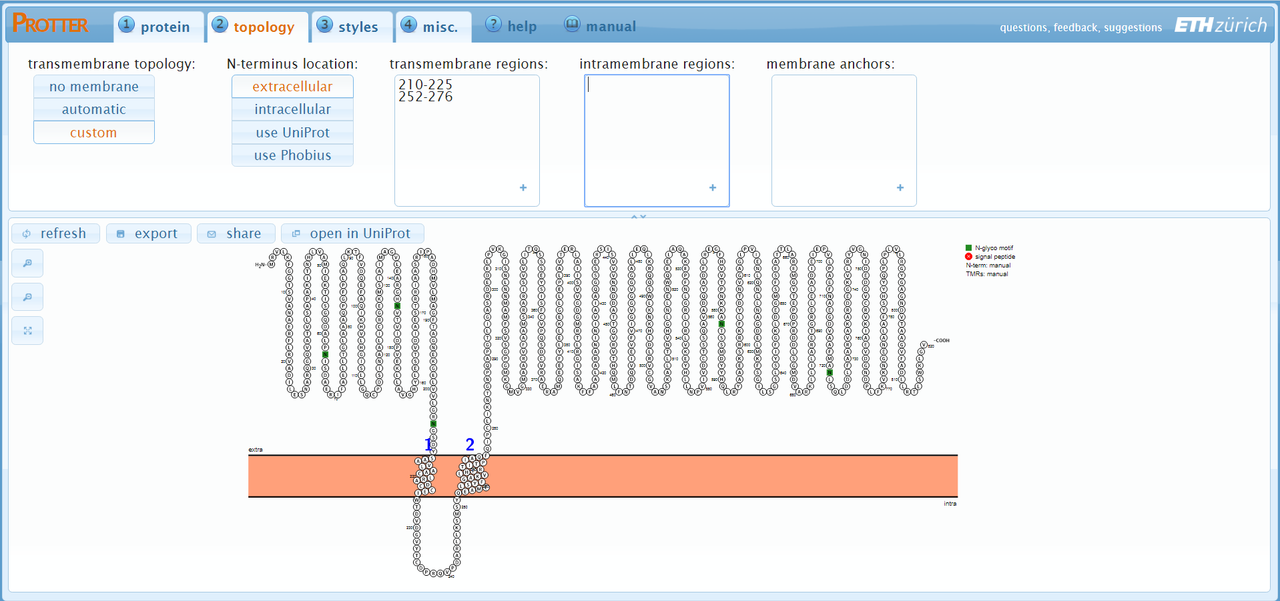
I have a transmembrane protein with unknown 3D structure.
I did membrane domain prediction on Protter and Phyre, but the pictures that the program generate are simple, and I need to create better pictures.
In that way I am trying to open the "flat structure" on PyMol or Chimera, but unsucessfull.
More specifically, I wanna create or reproduce a picture like the one figure 2A from the work DOI: 10.1126/science.aav0778.
Which is the input file format that I need to create to load on PyMol? How can I generate it?
Thanks in advance


Chimera and Pymol don't produce those types of images as far as I know (with the possible exception of some plugins), they're mainly for viewing full 3D crystal structures.
I have seen those diagrams before, but I'm not sure exactly what program makes them. You are closer with
Protter. You're looking for 'topology diagrams' essentially.Dear Joe, thanks for reply. I know chimera can produce it, by use "pipes and planks" toll. But I am not being able to generate a file that I can open at Chimera, as soon protter and phyre just generates a picture.
Phyre generates a PDB file. This is the input to Chimera. Protter doesn't do structural modelling, it appears only to use Uniprot IDs as far as I can tell.
You can use Phyre, SWISS-MODEL, ITASSER for instance, to generate a PDB file.
Other topology tools include:
But I don't know/think any of these do TM proteins specifically, they're generally for secondary structure tracing.
have you tried vmd? Not sure if this fits your interest, but this
is what I managed to make in vmd very quickly, I am sure one can work on it more to make it look more "pretty". Just in case this interests you : use the drawing method "Cartoon" in VMD
Dear manaswwm. Thanks for answer me. I did not tried vmd yet, but I believe the problem would be the same. The example that ou send me, are in 3D, but I do not have it. I just have the aminocids sequence, and the prediction from protter or phyre. So, I believe I would have the same problem with vmd, that is, which is the file format that I need to input. In other words, how to create a .pdb file ir other file that I can open at chimera/pymol/vmd, from a unknown 3D structure?
If you download VMD and go to File - New Molecule, then under Determine File type you can see all possible formats that VMD accepts, if this is what you mean. As for creating 3D structure from sequence information, you could perform homology modelling using SWISS-MODEL (which is easy to use web interface) or Modeller by SaliLab. Caution : models generated from homology modelling are just as reliable as your homologous protein's 3D structure
Dear all, Thanks for try to help, but I believe that I am not being able to explain exactly what I need.
I have the prediction, but what I mean, is that the protter or any other of those tools suggested generate "ugly" pictures. I need to prepare one more presentable, to add in a publication. Is more an esthetic problem, more than a bioinformatic.
For instance, from protter I have to adcy8 gene the picture at left, but I need something more like to the right one.
Can I generate a .pdb file from protter prediction and then, open it at Pymol?
You need to upload the image at something like imgbb and paste the HTML here. Yours hasn't rendered.
I'm still not exactly sure what you have Vs what you want.
Sorry for that. Hope it works:
Edited by @Joe to include images via direct HTML embedding.
Are you certain that the image on the right was generated by a tool? More often than not they're done by hand (which is annoying). If their methods/captions don't say exactly the tool that was used I'd advise getting in touch with the authors or the journal. If they provided Twitter contacts in the paper that's usually the quickest way.
Dear Joe. Thanks a lot. They say that all images of the article were made on Chimera or PyMol. But, yes, maybe this one was exception and they forgot to address. I write them. Thanks a lot.
Thats pretty common phraseology that tends to overlook some things indeed (or they simply fail to state how they made the images altogether). I'm willing to bet that wasn't made with PyMol or Chimera (though I'm less familiar with PyMol generally), but I have seen those style images enough that they could be generated by a tool I'm not aware of. Best indeed to just get in touch with the authors :)