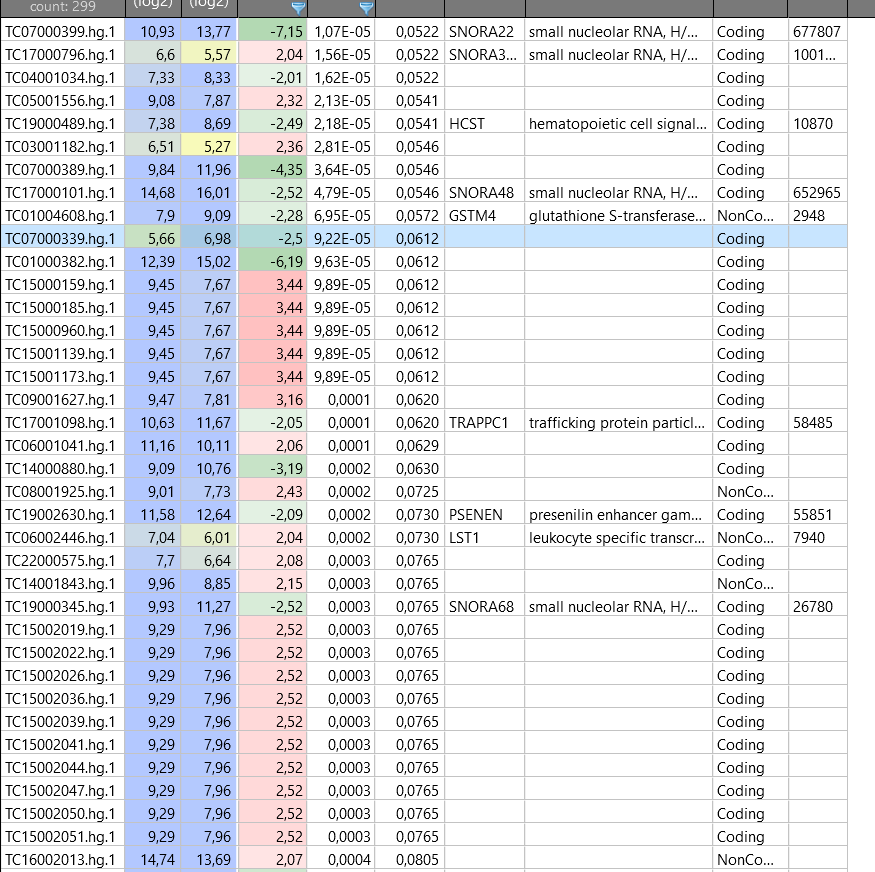
Some, or perhaps all, of these seem to relate to protein coding genes. The reason why they have no annotation could be because they were not yet assigned official gene symbols when this array was manufactured. Some of these already have Ensembl IDs, though.
There is extra information about them in the Net Affx file for this array: https://www.thermofisher.com/order/catalog/product/902162#/902162
grep -e 'TC07000339' HTA-2_0.na36.hg19.probeset.csv
"PSR07005056.hg.1","chr7","+","56123058","56123195","30","TC07000339.hg","Coding","EX07027581.hg","PSR07005056.hg.1","---","ENST00000383876 // chr7 // 100 // 0 // --- // 1 /// uc022adt.1 // chr7 // 100 // 0 // --- // 1","main","1","TR07002358.hg","ENS//UCG","Constituitive","1","100.00 %","pass","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---"
grep -e 'TC15002026' HTA-2_0.na36.hg19.probeset.csv
"PSR15018739.hg.1","chr15","-","102294519","102294548","6","TC15002026.hg","Coding","EX15013431.hg","PSR15018739.hg.1","---","DQ575740 // chr15 // 100 // 0 // --- // 1 /// DQ575740 // chr15 // 100 // 0 // --- // 1 /// uc021syd.1 // chr15 // 100 // 0 // --- // 1","main","1","TR15007323.hg","UCG","Constituitive","1","100.00 %","pass","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---","---"
I would download the Net Afffx file and find some way to add the annotation. As you are using the Affymetrix Expression console (?), there must be a way to add extra fields from the Net Affx file. Otherwise, you could simply perform the analysis on the command line, which would be easier in many respects.
Kevin


Can you confirm the input files that you used? For example, which NetAffx file?
I used GeneChip™ Human Transcriptome Array 2.0 microarrays. The raw files are .CEL files