I am wondering how a single ionized peptide can produce fragments offset by one amino acid, as shown in this figure:
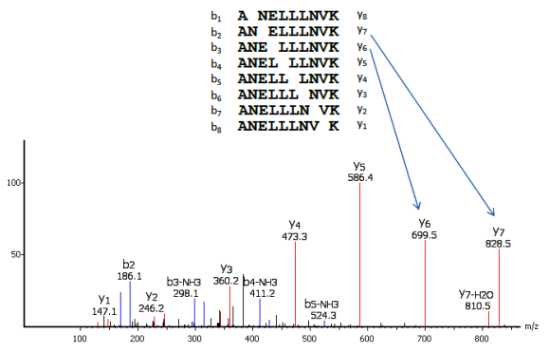
For instance, if your peptide is ANELLLNVK, I could understand producing fragments ANEL and LLNVK for example, but how A, AN, ANE, ANEL etc? How can a residue be contained in another fragment if already encompassed in one?
This all started because I was wondering how mass spec proteomics can produce the correct directionality of a peptide sequence. It must be because each residue adds a known mass, thereby increasing the m/z and yielding the full peptide sequence.
This has been bugging me for hours and I must be missing something!

