Hello! I'm creating PCA plots with a whole transcriptome dataset, which includes 26,828 'genes', to assess similarity between three groups (E, M and L). I'm doing differential expression analysis using DESeq, so I transformed my DESeqDataSet using VST, then used the plotPCA function from this package, with the following code:
library(DESeq)
plotPCA(vsd, intgroup = c("dev"), ntop = 26828)
This produced this PCA plot, with 78% variance in PC1 and 14% in PC2:
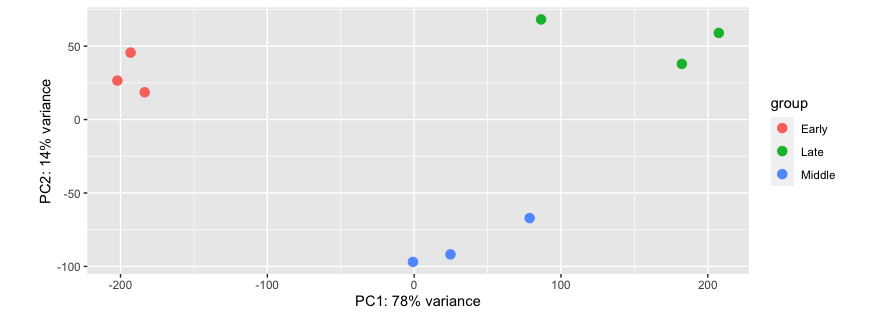
I'm not sure how to upload my transformed DESeqDataSet in an accessible format for trouble shooting. However, the above plotPCA function uses the stats package prcomp function to produce the same plot, with the following code:
pca<- prcomp(vsd.transposed)
pca$x %>%
as.data.frame %>%
ggplot(aes(x=PC1,y=PC2)) + geom_point(size=3) +
theme_bw(base_size=32) +
labs(x=paste0("PC1: ", percentVar[1], "% variance"),
y=paste0("PC2: ", percentVar[2], "% variance")) +
theme(legend.position="top")
Subsequently,I wanted to look at the genes and GO terms correlated to PC1 and PC2, which is possible using dimdesc in FactoMineR. I created another PCA plot of the same data using the FactoMineR function PCA.
library(FactoMineR)
FactoMine.pca <- PCA(vsd.transposed, graph = F)
plot((FactoMine.pca), axes=c(1,2))
This plot looks fairly similar to the first one, but the proportion of variances explained by Dim 1 and 2 are quite different compared to the plot produced by plotPCA.
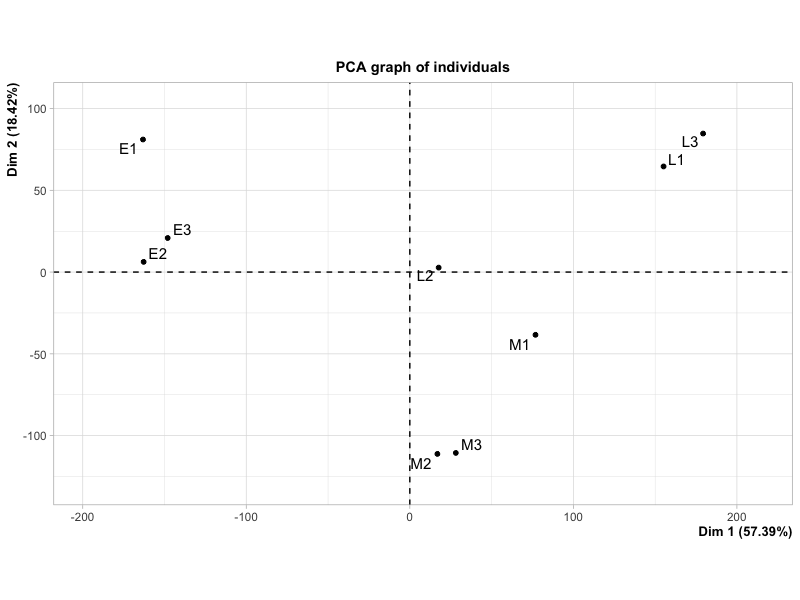
Is this difference due to an error in my code or the computing method used by the two different functions? If this is the case, is there one which is generally preferred?
Thanks for your attention!
EDIT: Thank you for your help everyone - the difference was due to the use of scaling by default in FactoMineR but no scaling in plotPCA!



Please see How to add images to a Biostars post to add your images properly. You need the direct link to the image, not the link to the webpage that has the image embedded (which is what you have used here)
Also, post your code/data on a GitHub Gist and paste the link to the gist here (See how-to post here). Don't use sites like ufile and force other users to download files just to help you. Remember, we're not customer support.
Thanks for the information, I have updated the post accordingly!