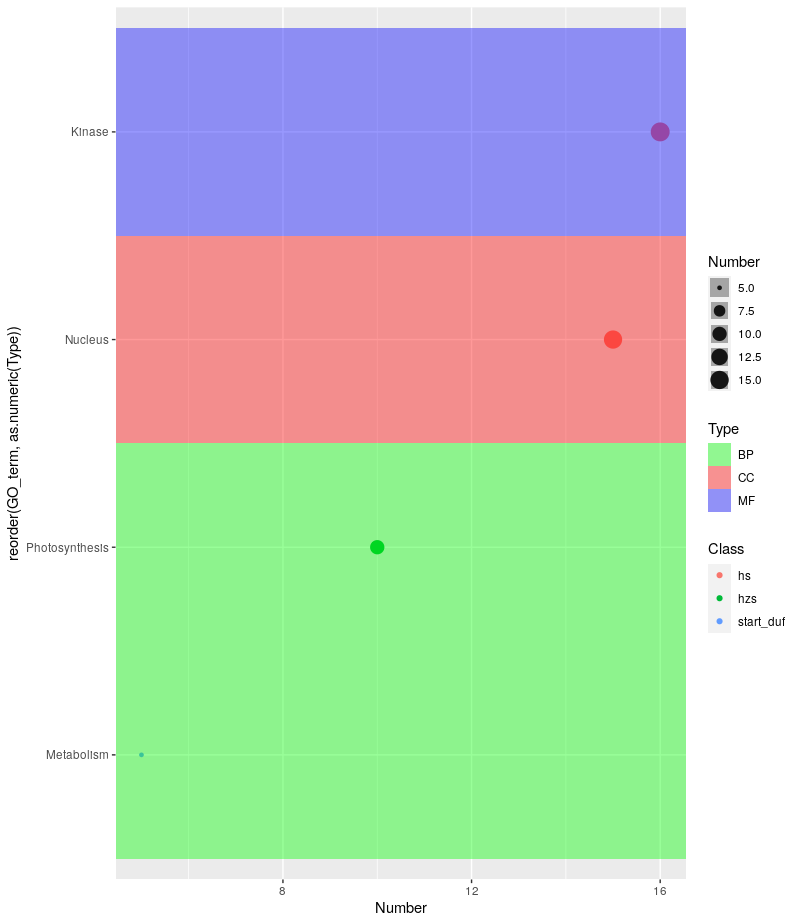
Dear all, I want to ask a very basic question. I am making a bubble plot using ggplot2 having table structure:
GO term Number Class Type
1. Metabolism 5 start duf BP
2. Photosynthesis 10 hzs BP
3. Nucleus 15 hs CC
4. Kinase 16 hs MF
I want to make a bubble plot having Number on x axis, GO term on y axis, bubble color should be based on Class and background color will be based on Type. My R code is:
ggplot(bubble_plot, aes(x=Number, y=GO term, size = Number, col = Class)) + geom_point(alpha=0.7)
With this I am getting the desired plot except the background color, When I am using "fill = Type", I am not getting the background color based on the BP, CC or MF based on the " Type" column in the table.
The desired plot should look like:
Please help.
Thanks in advance



Awesome! It worked perfectly.
Thanks for your help.
Cheers
Please accept the answer (green check) mark to provide closure to this thread.
Hi, Sorry to disturb you again. I am getting the desired result with your code but getting some strips darker than others. Is there any way to keep it uniform? I am getting something:
Hi, no worries! Could you please perhaps share your code with me? It looks like that transparency for the tiles (
alpha) is being set conditionally? It could also be because of theTypevariable (I'm not sure what that's being passed as toggplot()).Thanks for your response. I am using the same code that you have mentioned:
ggplot(mydat, aes(y = reorder(GO_term, as.numeric(Type)), x = Number, size = Number)) + geom_point(aes(color = Class), alpha = 3.0) + geom_tile(aes(width = Inf, fill = Type), alpha = 0.2) + scale_fill_manual(values = c("green", "red", "blue"))
When I am reducing alpha in geom_tile, I am getting some strips darker than others.
Thanks again
Hmm I think I know what's happening. Since every point on the plot is getting its own
geom_tile(), the colors are darker in cases where there is more than one point in the same row (because successive tiles of the same color are being overlaid one on top of the other). I didn't realize that would happen because my little toy dataset did not have datapoints that fell in the same Y-axis "row".So I have a workaround for you. I'll use my original toy example, modified with an additional point in the
Photosynthesisrow to illustrate how this works. Basically what we'll do is create a new column calledtypefillthat will be used to set thefillforgeom_tile(). Since each "row" can have overlappinggeom_tile()s we are going to set the value intypefillconditionally. The condition is this: we will group all the rows (of thedata.frame()) together that have the sameGO_term(Y-axis value) andType(our originalgeom_tile()fillvalue). Now for each of these groups of rows, we assign theTypevalue of the group as thetypefillvalue of the first row of that group; all other rows just get anNA. The result is that when we plot the data now, and passtypefilltogeom_tile()'sfillparameter, we will no longer have overlaid colors as we saw in your output as the fill is being set only once.And this is the result: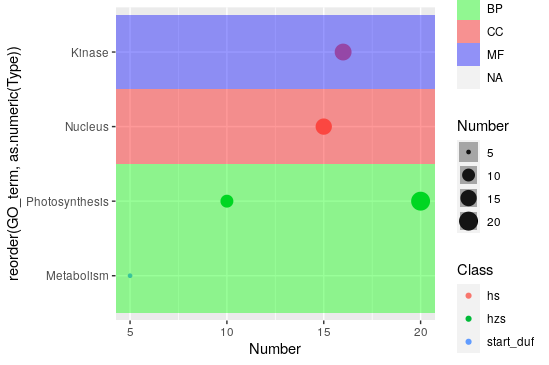
Of course, you now have that one extra
NAcolumn in the legend, but that can be hidden quite easily.Great! Thanks. It now works perfectly. Sorry to disturb you so much.
Thanks again
Oh not at all, I am glad I could help. Don't hesitate to ask if something goes sideways again!!
Hello Dunois
I would really appreciate if you help me in reordering the y-axis using reorder function. I used the same code as you suggested but I am unable to replicate it. Please see this link: enter link description here
Hello
What is 'Class" in your tabular data? Is it specific to your data or it's some kind of general representation in GO classification? My data only has GO terms, count, and GO category?