I am starting in the field of bioinformatics, could you indicate good metagenome assemblers?
1
Entering edit mode
3.6 years ago
Mensur Dlakic
★
27k
Don't know how one can choose the best assembler, but these two should be a good starting point:
0
Entering edit mode
3.1 years ago
juanjo75es
▴
130
Actually, if you are dealing with viruses or phages these are the best assemblers compared using a public benchmark set:
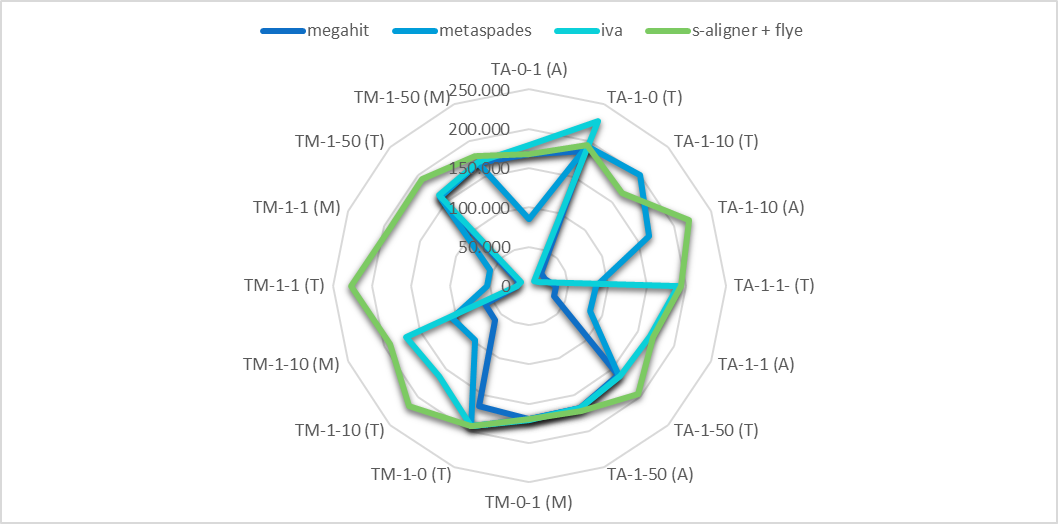
s-aligner is first followed by metaSPAdes.
Reference: s-aligner: a greedy algorithm for non-greedy de novo genome assembly
0
Entering edit mode
3.1 years ago
956120746
•
0
Hi: After the test in our lab, mitofinder --metaspades gain the best result, you can also get mitochondrial data.
Similar Posts
Loading Similar Posts
Traffic: 2955 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Please read:
(short version) - [[ Please read before posting a question ::: How To Ask A Good Question ]]
(long version) - How To Ask Good Questions On Technical And Scientific Forums
Then edit your post to include more information, such as the what type of data you have, how much data, and so on. Also, perform a good bibliographic review and see which assemblers would do a good job for your data.