Hello,
I'm practically a newcomer to bioinformatics and I'm a bit stuck with a thing. We're working on DNA repair/recombination and we are currently sequencing a particular region to analyse recombination inside it in plant hybrids. So basically I have Illumina seq data of PCR amplicons of the region (450bp, so each paired-end read covers the whole region) that I aligned and called haplotypes with freebayes to have a vcf with a set of SNPs determining the two haplotypes in the region of both the parental backgrounds of my hybrid. Now I would like to re-run my BAM files against the known haplotypes in order to call the fraction of reads that match to one parental (ej. AAAA), to the other (BBBB) and the different recombinants (ABBB, AABB, AAAB...). But I cannot find what tool to use to get the metrics and output also different BAM files for each parental and recombinant class if possible.
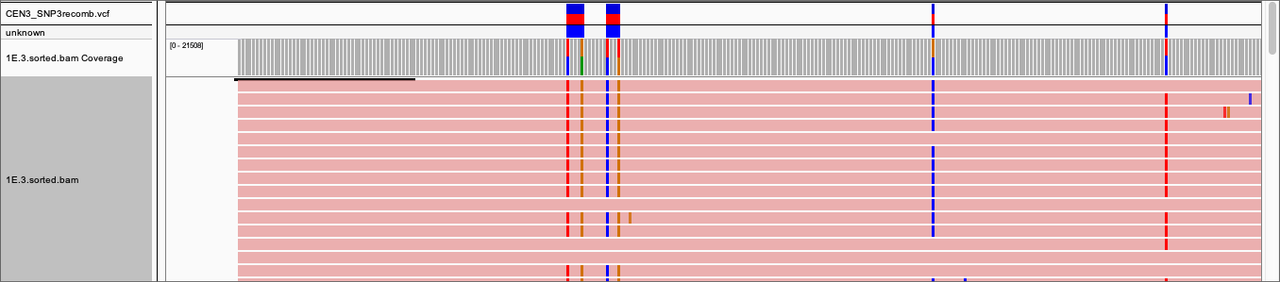
I include a panel of IGV with the result of haplotype calling on top and some reads as example. They are kind of obvious to count visually, but I would like to learn how to do it with some tool as we are about to scale to high numbers and more complex/longer regions.