Entering edit mode
3.1 years ago
baliczap
•
0
Dear All,
I frequently see such variant on genome browser when looking at variants. I was wondering what is the source of this? It seems as it would be a haplotype, but quality is frequently low by variants such as this. I thought on pseudogene effect, but could not prove so. Any idea?
Thank you
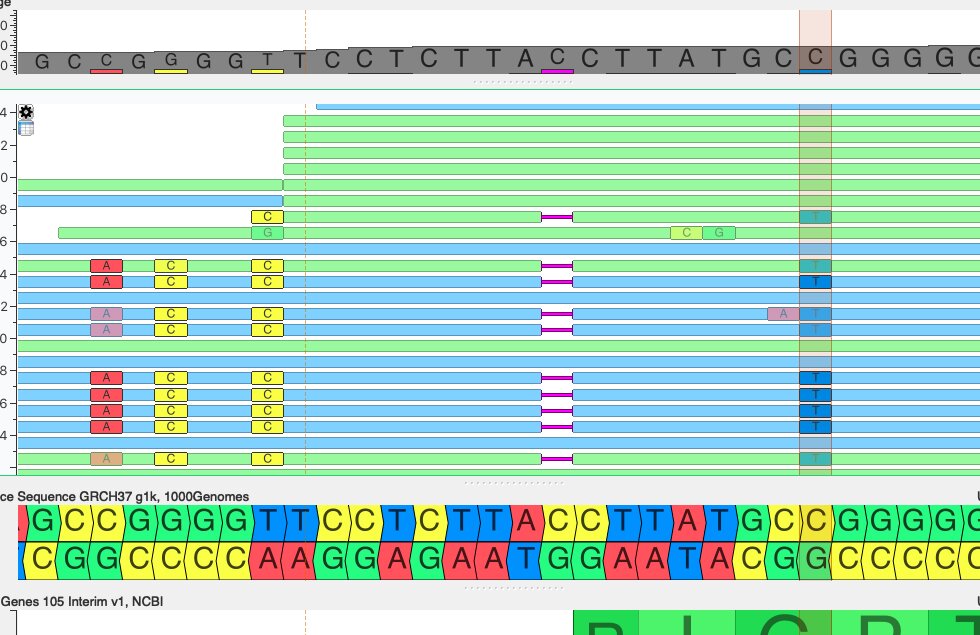


Hi,
These kind of variants are usually called delins and there are some pathogenic known delins variants. In this article there are some suggestions of possible causes of such mutations: