I ran FatsQC on a set of 4 Illumina PE exomes and got really weird graphs for the kmer distribution. I've never seen this pattern before - does anyone know what is going on and what to do about it?
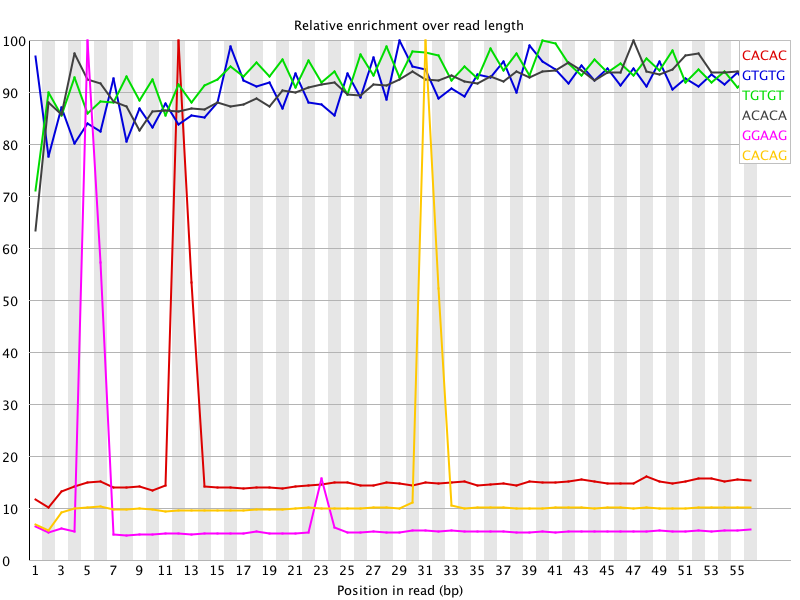
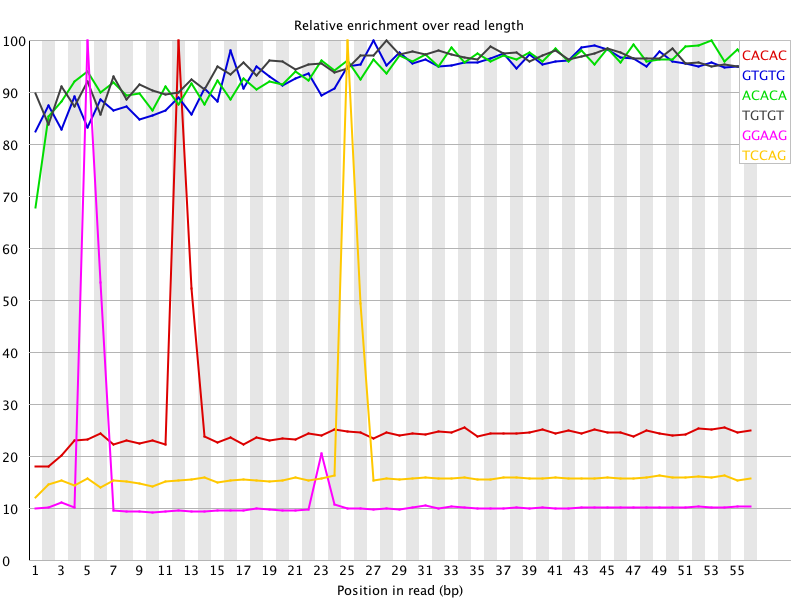

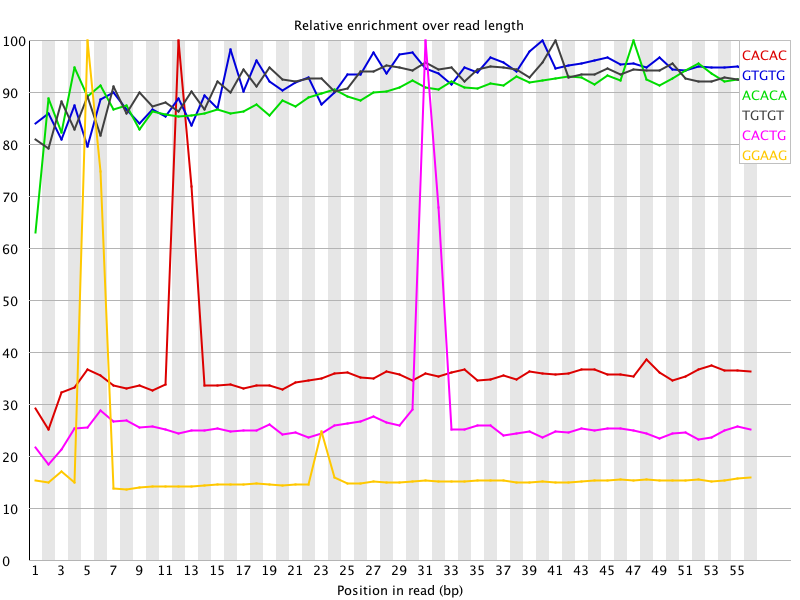
I ran FatsQC on a set of 4 Illumina PE exomes and got really weird graphs for the kmer distribution. I've never seen this pattern before - does anyone know what is going on and what to do about it?




Those k-mers almost perfectly correspond to the TruSeq adapter sequence. Was there anything peculiar that showed up in the overrepresented sequences table?
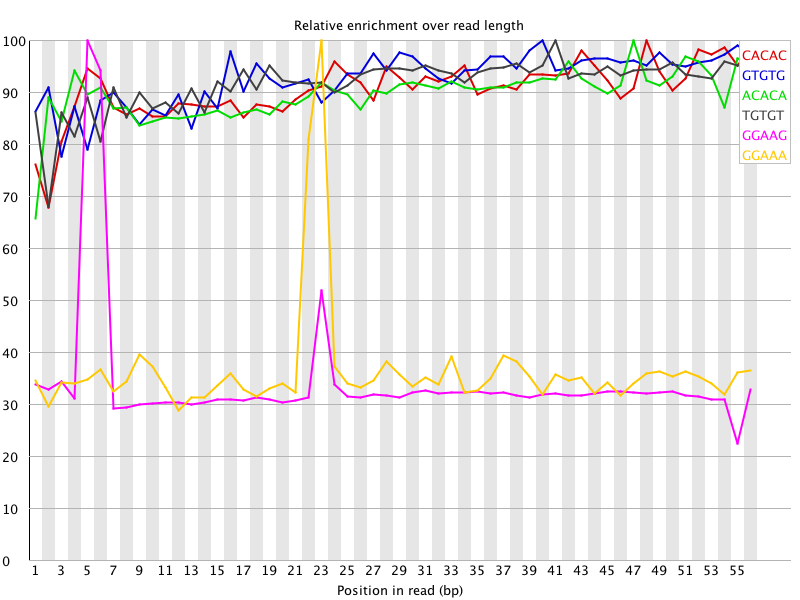
@kelvin.c.chan: I did remove the over-represented sequences for one of the samples and the graph changed somewhat but there are still peaks (see below) although fastqc no longer reports any over-represented sequences. Any ideas?
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.
You are right - there are TruSeq adaptors among the overrepresented sequences. I'll remove them and then re-run fastqc.