All practicing bioinformaticians will face problems that require them to compare, query and select genomic features across an entire genome. As it happens efficient interval representation and query is a surprisingly challenging problem that needs a specialized representation.
The BEDTools suite contains a set of programs that support a broad range of interval analyses that involve selecting certain locations in the genome. The name reflects the original intent to process BED files but the tools operate just as well on GFF formats. The scripts need to be run in command line format and are available for UNIX type systems: Linux, Mac OSX, and Cygwin (on Windows).
The link to the site is: http://code.google.com/p/bedtools/
With BEDTools one can answer questions such as:
- how many reads map upstream/downstream of one or more locations in the genome?
- how many reads cover a certain base in the genome?
- which sections of the genome are not overlapping with target intervals?
- what are the sequences specified by the coordinates?
- ...
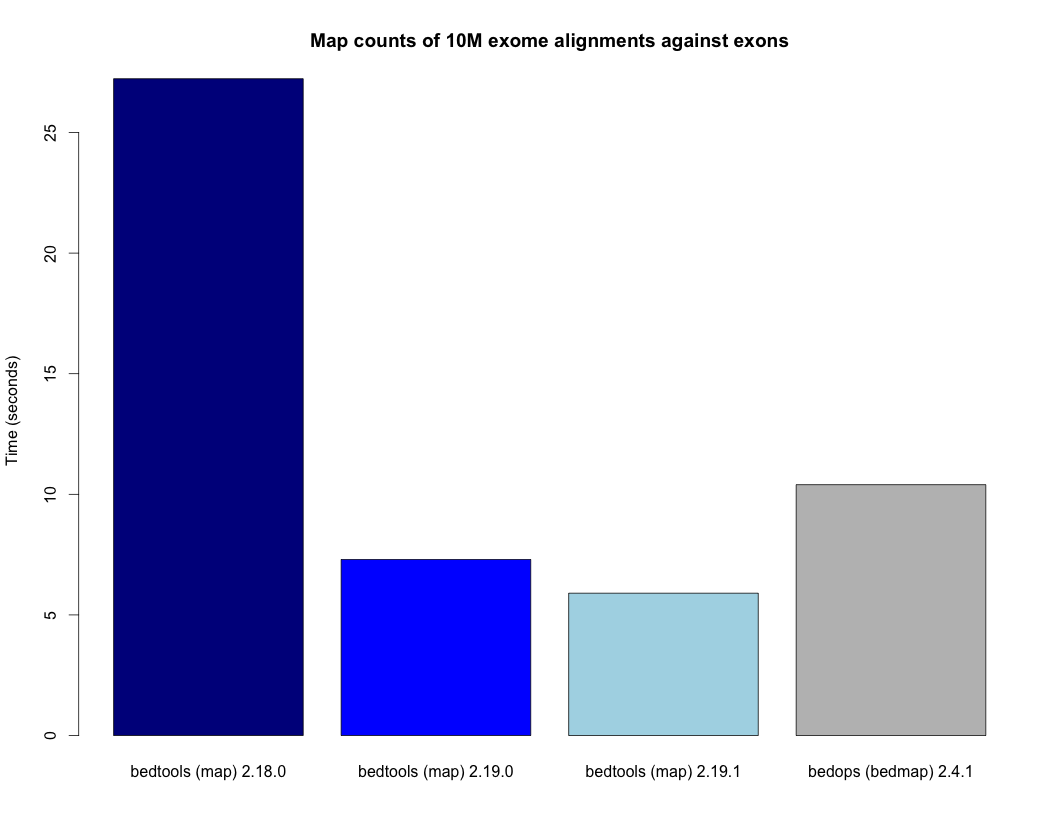
The suite consists of multiple tools but for beginners the most important is intersectBed. Understanding this tool is a gateway to understanding them all. In fact many (but not all) of the other tools slopBed, windowBed are simply convenience tools that assist users preparing/formatting output a certain way and could be replaced by small custom scripts.
Note: a very large number of problems can be solved via running nothing more than the various scripts in BEDTools and occasional reformatting of the outputs. If you are new to the field take your time and learn what BEDTools does.


Can you post the bedtools recipe for annotation of intervals by features such as TSS CDS Exons 5' UTR Exons 3' UTR Exons CpG Islands Repeats Introns Intergenic