Hello,
I have created a Blast database using a reference genome. Then, I have performed a local blast search in command line using a gene of interest. I have obtained some hits with the usual Blasting information. Now, I want to extract the exact matching sequence from the blast search with their corresponding start and stop position. I have a .gff file for the genes as well for the reference genome.
Can you give me any suggestions on how to do that?
Thanks a lot.
If you want to stay in the command line you can:
- run blast with the text/table output format. This gives you the identifier, start and stop locations for each hit (among other things)
- using the hit identifier column (the 2nd I believe), extract the FASTA from the blast database using
blastdbcmdas highlighted by @genomax. Alternatively, extract the fasta sequence directly from the original reference genome FASTA (genome tools can help you extract just a specific subsequence).
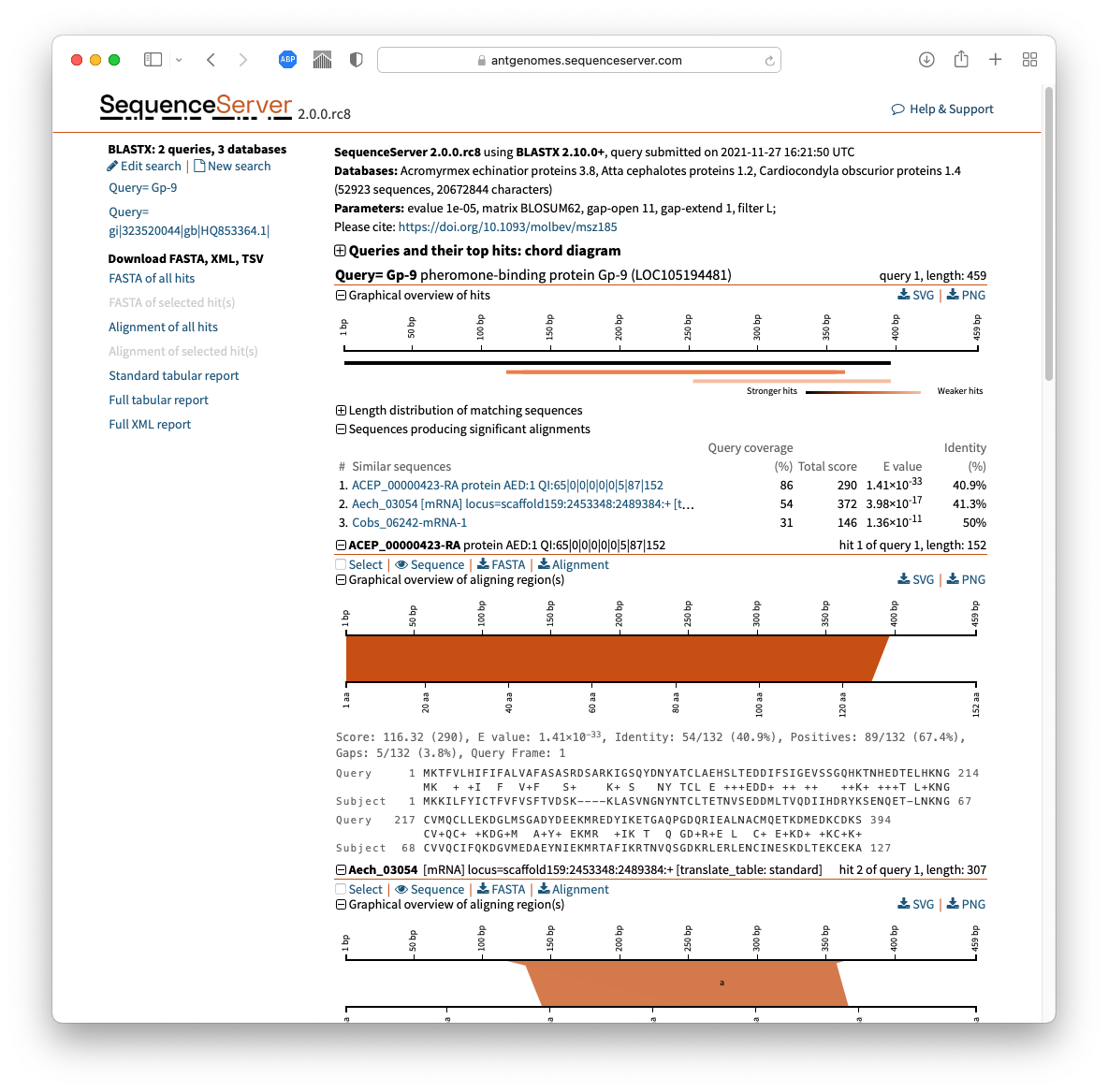
Or if you prefer a point-and-click interface, use SequenceServer to run your BLAST search (FYI - my lab makes the tool; we're proud of it; it's open source). Then in the Download FASTA, XML, TSV section on the left side of the screen (see screenshot below)
- click on "FASTA of all hits"
- OR if you just want the aligning segments, click "Alignment of all hits" and retain only the hit sequences from those pairwise alignments.
- OR select just some of the hits using the [ ] checkboxes & download their entire sequences or aligning parts
The screenshot below shows the download buttons on the left - and shows how next to each hit is a selection checkbox
Yannick
Thank you so much for an elaborate reply. Actually, I want to stay on command line and build a pipeline where I can easily blast a gene sequence against a reference and then extract the sequence information from other samples. Right now, I have created a local blast database using the reference genome of interest and also could blast a gene of interest. I was thinking if there is any chance to extract the blast sequence and get their coordinates from the .gff file within command line. What you showed is quite nice but I was thinking of automating these steps within command line. Can you give some feedback on that please?
I think I still don't understand exactly what you're trying to do. bedtools will likely be your friend - if you get the right columns of the BLAST table output, its effectively a bed file. You can use bedtools to find overlaps between this new .bed file and your existing .gff
Yes, exactly. That is what I just performed. What I want to do is:
- Create a local blast database with the reference genome of interest
- Blast the gene of interest against the reference genome
- Use the best hit and extract the Start and Stop Position
- So you have an interval file
- Use that interval file to extract variants from the VCF files of different samples.
Sorry, I should have made it clear before.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Use
blastdbcmd: https://www.ncbi.nlm.nih.gov/books/NBK569853/thank you for the useful link