Minigraph as a multi-assembly SV caller
Honestly, I didn’t know what minigraph would be good for when I was writing the code. When I was writing the paper, I pitched minigraph as a fast caller for structural variations (SVs). However, except performance and convenience, minigraph is not that special. In fact, in the paper, minigraph is not as good as read-based SV callers because it randomly misses one parental allele when most assemblies in the paper are not phased.
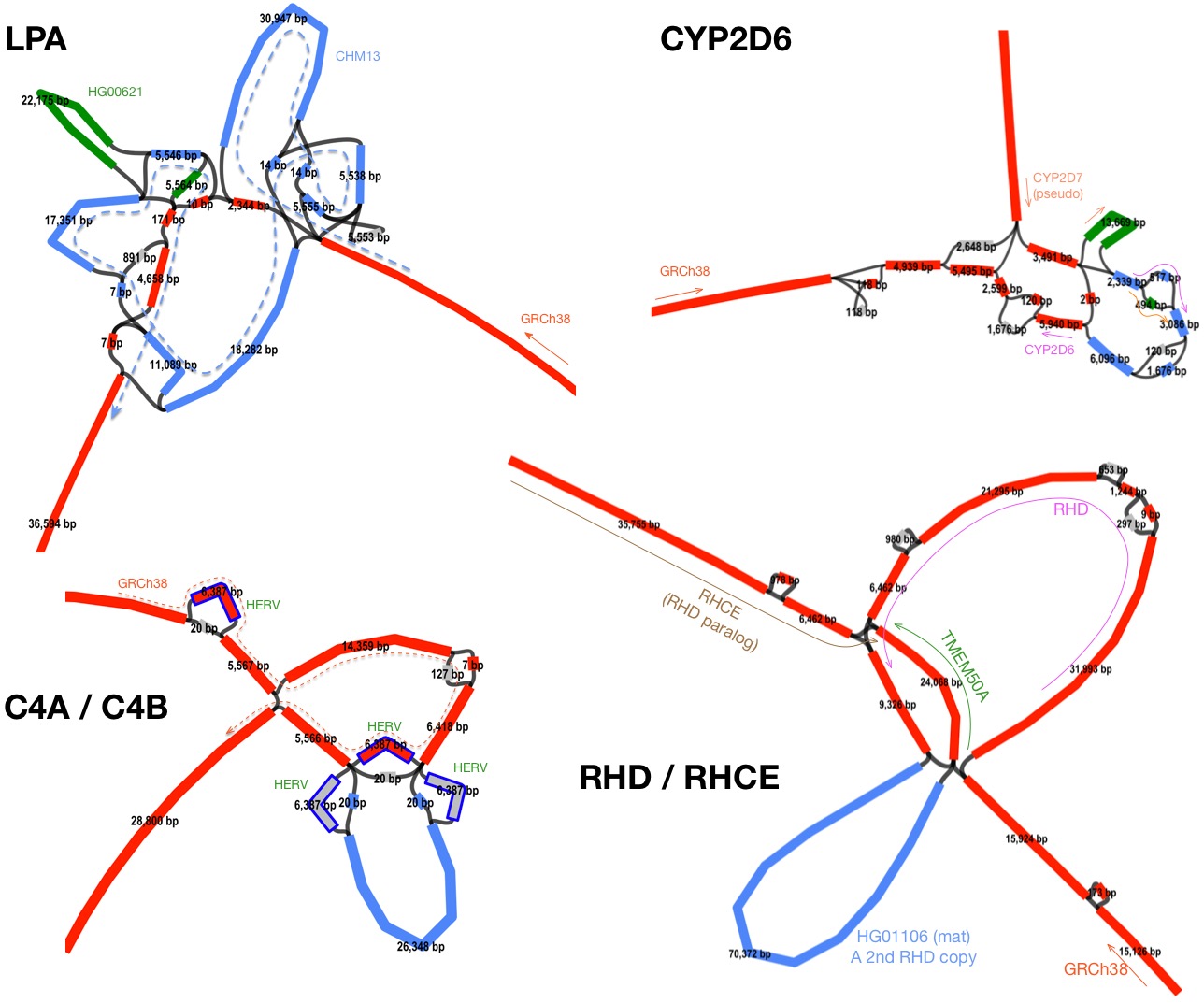
My exploration took a turn when one anonymous reviewer asked me to check the LPA gene. It was not in the graph because the gene was collapsed or missed in all input assemblies. Fortunately, I had several phased hifiasm assemblies at hand. LPA is there and minigraph generates a complex subgraph (figure below) far beyond the capability of VCF. Then I realized what minigraph is truly good for: complex SVs.
With the current SV calling pipelines, we typically map reads or an assembly against a reference genome, call SVs and then merge pairwise SV calls into a multi-sample call set. This sounds simple but doesn’t work well for complex events. First, the position of an SV may be shifted by small variants. We have to heuristically group nearby events. This is particularly problematic around VNTRs. Second, there are nested SVs: for example, an L1 insertion inside a long segmental duplication. If we only see the reference coordinate, we wouldn’t be able to easily represent duplications with and without L1.
The solution to these problems is multi-sequence alignment (MSA) which minigraph approximates. MSA naturally alleviates imprecise breakpoints because MSA effectively groups similar events first; MSA also fully represents nested events because unlike mapping against a reference genome, MSA aligns inserted sequences not in the reference. The following figure shows the subgraphs around four genes. SVs like these will fail most existing SV callers and can’t be represented in VCF.
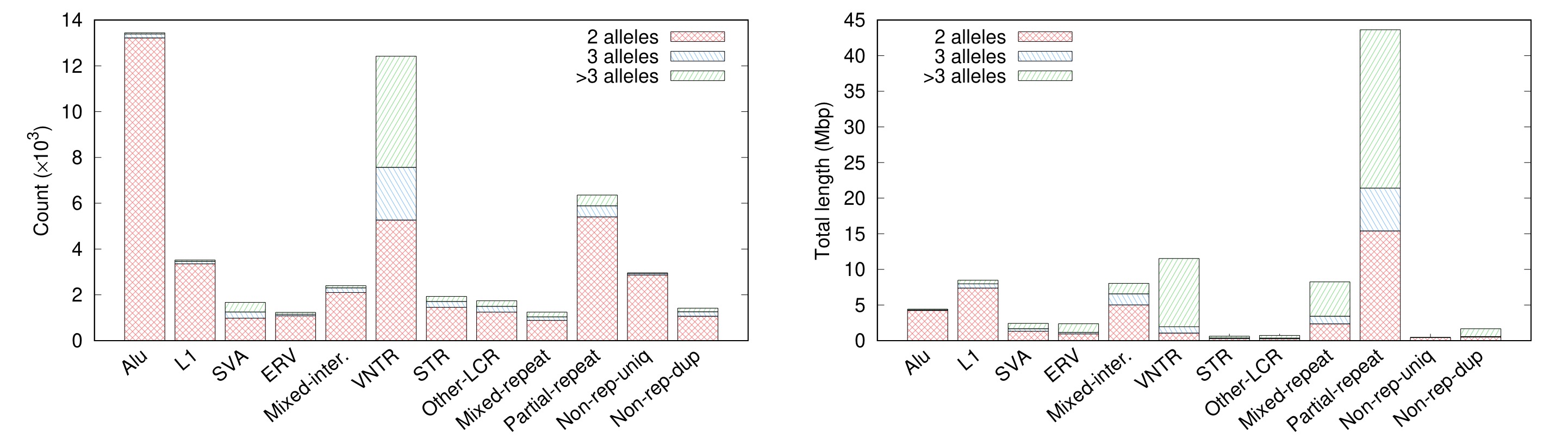
Are there many complex SVs? Not a lot by count. In the left plot below, all examples come from blue and green areas on the “Partial-repeat” bar. There are only several hundred of them. However, these complex SVs often reside in long segmental duplications and affect a much larger fraction of genomes in comparison to transposon insertions (the “Partial-repeat” bar on the right plot). Genes in these loci, a few hundred of them, are frequently related to immune systems (e.g. many HLA/KIR genes) or under rapid evolution in the primate or human lineage (e.g. AMY* and NBPF* genes). My last blog post mentioned 10% genes that have multiple copies in CHM13 are single-copy in GRCh38. These genes mostly come from the “Partial-repeat” bar, too. With short reads, we can observe signals of transposon insertions and copy number changes and with long reads, we can call VNTRs, but only with multi-assembly callers like minigraph, we can have the access to the near full spectrum of SVs, with the exception of centromeric repeats.
Minigraph is a fast and powerful multi-assembly SV caller. Although the calling is graph based, you can ignore the graph structure and focus on SVs only. I have just added a new section in README that explains how to use minigraph to call SVs. It is worth noting that at complex loci, minigraph subgraphs, including examples above, are often suboptimal. Please read the Limitations section if you want to explore the minigraph approach.
blog comments powered by Disqus