DEE2 projects on demand
We have noted that the time between new datasets appearing on SRA and being processed by DEE2 has been about 3 to 6 months. Our dream is to shrink this down to two weeks, but we simply do not have access to that much compute power at the moment. To address this we have devised an "on-demand" feature so that you can request certain datasets to be processed rapidly. We think this is a great feature because it serves the main mission of the DEE2 project which is to make all RNA-seq data freely available to everyone.
Here's how to use it:
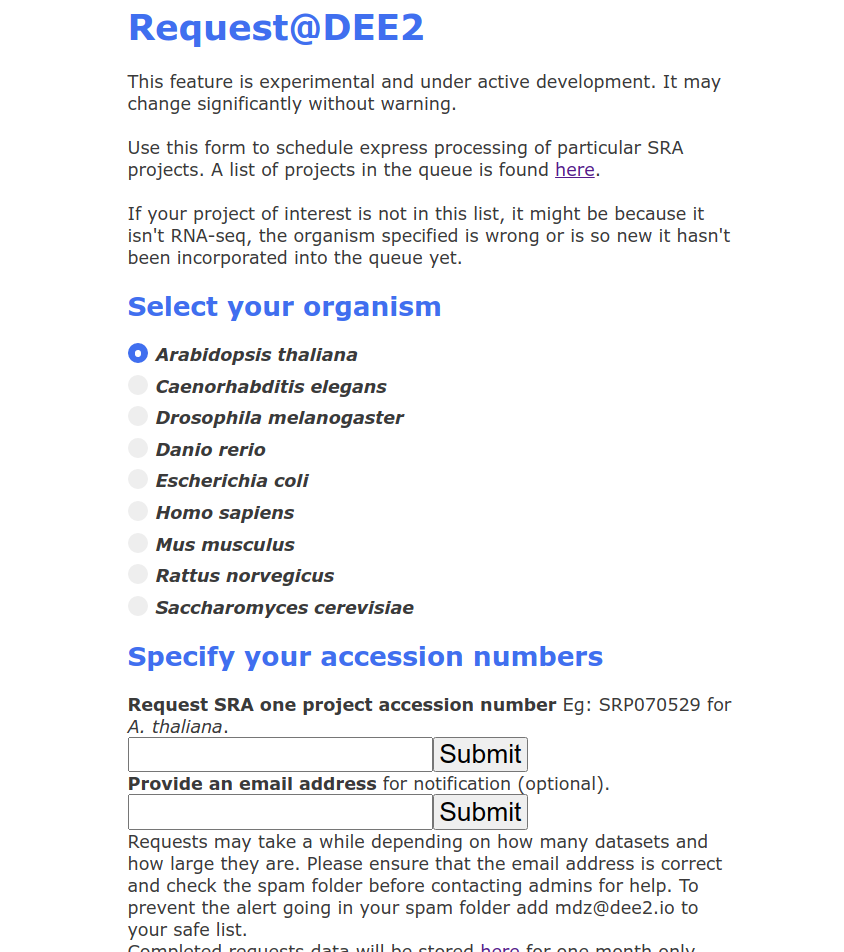
1. Visit http://dee2.io/request.html and you will be greeted with a webform. Select the organism of interest.
2. Provide the SRA project accession number of the dataset. These numbers begin in SRP/ERP/DRP. If you have a different type of accession such as GEO Series (GSE) or Bioproject (PRJNA) then you will need to navigate NCBI to find the SRP number.
3. Check that the SRP number is in the standard DEE2 queue. To do that, follow the link provided above the request web form, click on the queue that corresponds to the organism of interest and use ctrl+f to search the text file.
4. If it's there, return to the webform and paste or type the SRP accession into the first text box. Give your email address as well if you want to be notified that processing has completed.
5. You will receive an email when processing is complete. The email will indicate whether any errors were encountered. There will be a link to a folder of recently requested datasets here.
6. Download and enjoy the dataset as you would any DEE2 data.
Video walkthrough
Boring technical details
Here how it works:
* There is a html webform which executes a cgi script and writes to a file on the webserver that stores the request. Each request consists of a species, SRP number and optional email address.
* Every 10 minutes, a remote computer runs a script which downloads the request file and checks to see if there are any new requests. If there are, it processes all runs in the SRA project with the docker pipeline.
* After processing, the data files are copied out of the docker container, tabulated into matrix format for downstream analysis, zipped and uploaded to the webserver.
* Every 5 minutes, the webserver runs a separate script which checks whether requested datasets have been completed and present in the folder. If it is, then an automated email is sent to the user to notify the data is ready.
This efficient approach ensures that users can rapidly access new and interesting datasets, and that these data are incorporated into the monthly updates. If this becomes popular then there is definitely scope to increase the available compute power.
I would love to hear your feedback and suggestions.