Entering edit mode
13 days ago
Emmi
•
0
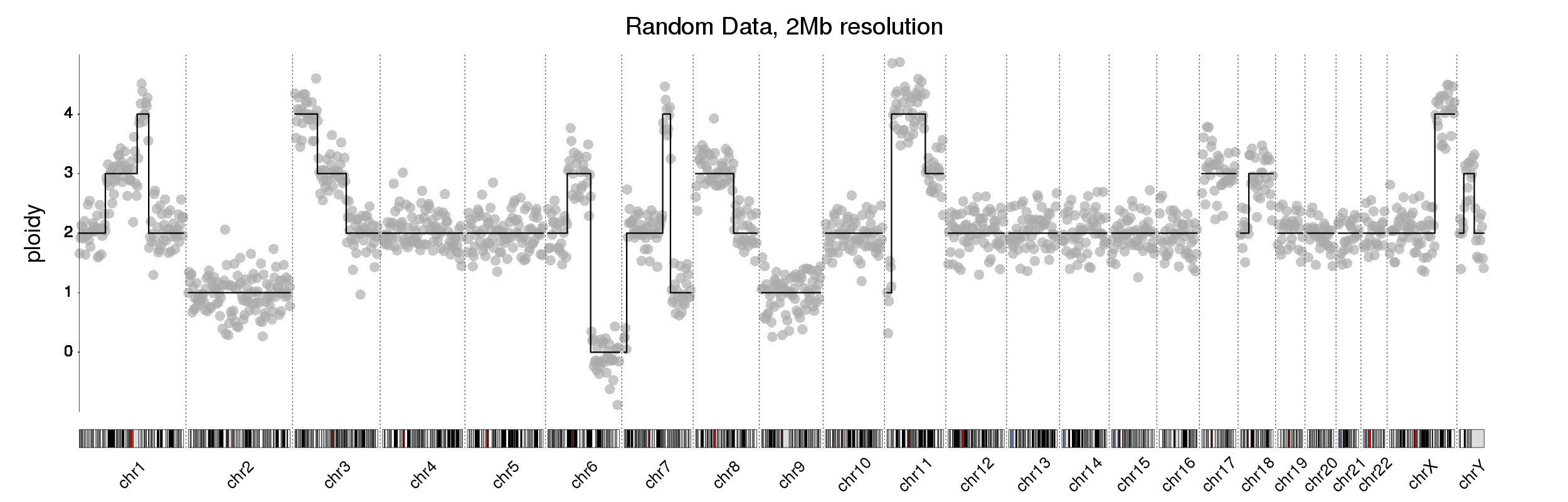
Hi guys,
I am looking to create a plot like this. There has been a previous post on the forum on this with the plot from @bernatgel. I loved the solution but am unable to translate it to my data. Maybe someone knows?
I am looking to create this plot using karyoploteR.
My dataset looks like this (with made up examples below):
- chromosome number
- start: denotes start position of copy number variation
- end: denotes the end position of the variation on the chromosome
- ploidy
log2_ratio
chromosome start end ploidy 1 58865614 58865899 3.512352 2 14538784873 14538784998 3.512352 4 83948882 839497809 3.512352 4 82748427 827486789 3.512352
How do I adapt the code below, to work on my dataset?
kp <- plotKaryotype("hg19", plot.type = 4, labels.plotter = NULL, main="Random Data, 2Mb resolution", cex=3.2)
kpAddChromosomeNames(kp, srt=45, cex=2.2)
plotLRR(kp, tiles, ymin=-1, ymax=5, labels = NA, points.col = "#AAAAAAAA", line.at.0 = FALSE, points.cex = 3, add.axis = FALSE)
plotCopyNumberCallsAsLines(kp, cn.calls = rr, ymin=-1, ymax=5, lwd=3, add.axis=FALSE, labels = NA)
kpAddChromosomeSeparators(kp, lwd=2, col = "#666666")
kpAxis(kp, ymin = -1, ymax=5, tick.pos = 0:4, cex=2.2)
kpAddLabels(kp, labels = "ploidy", cex=3, srt=90, pos=3, label.margin = 0.025)
Help would be greatly appreciated!

