Entering edit mode
5.9 years ago
gundalav
▴
380
I have the following code:
library(Gviz)
library(Mus.musculus)
library(data.table)
library(OrganismDbi)
library(TxDb.Mmusculus.UCSC.mm10.knownGene)
txdb <- TxDb.Mmusculus.UCSC.mm10.knownGene
# Define bam file -----
adipose_bam <- "myfile.bam"
# Define region -----------------------------------------------------------
thechr <- "chr3"
st <- 116.108e6L
en <- 116.1317e6L
# Define track ------------------------------------------------------------
itrack <- IdeogramTrack(
genome = "mm10", chromosome = thechr
)
grtrack <- GeneRegionTrack(
txdb,
chromosome = thechr, start = st, end = en,
showId = TRUE,
name = "Gene Annotation"
)
gtrack <- GenomeAxisTrack(cex = 1) # set the font size larger
adipose_altrack <- AlignmentsTrack(
adipose_bam,
isPaired = TRUE, col.mates = "deeppink",
name = "Adipose", fill = "blue", col = "blue"
)
plotTracks(
list(itrack, gtrack, adipose_altrack, grtrack),
from = st, to = en
)
It produces this plot: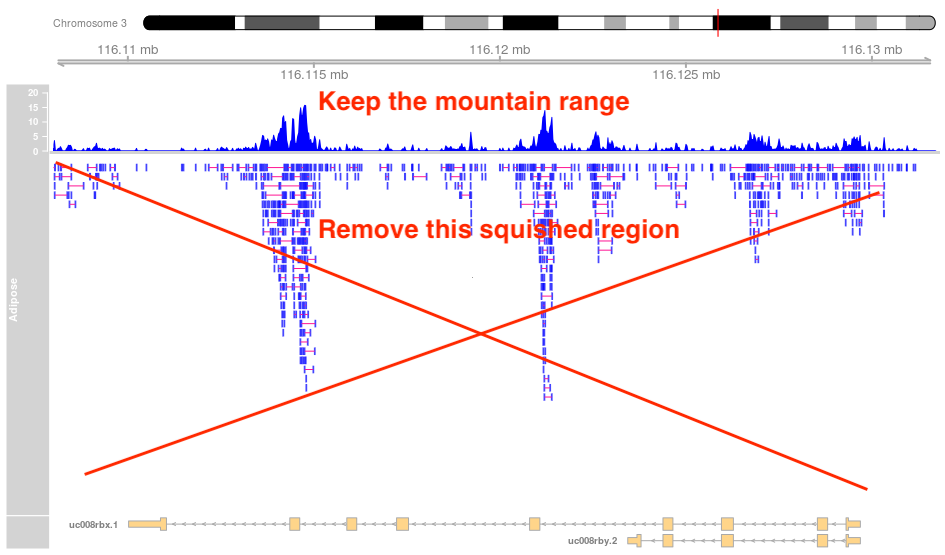
As define in the image how can I remove the squished track?

