Hi and sorry for the long post in advance,
I'm doing a survival analysis of lung cancer patients using Python's lifelines package. According to the documentation, the function plot_partial_effects_on_outcome() plots the effect of a covariate on the observer's survival. I do understand that the CoxPH model assumes that the log-hazard of an individual is modelled by a linear function of their covariates, however, in some cases the effect of these covariates seems to perfect when plotted. I'll illustrate these with three images below describing the issues I have in my data analysis.
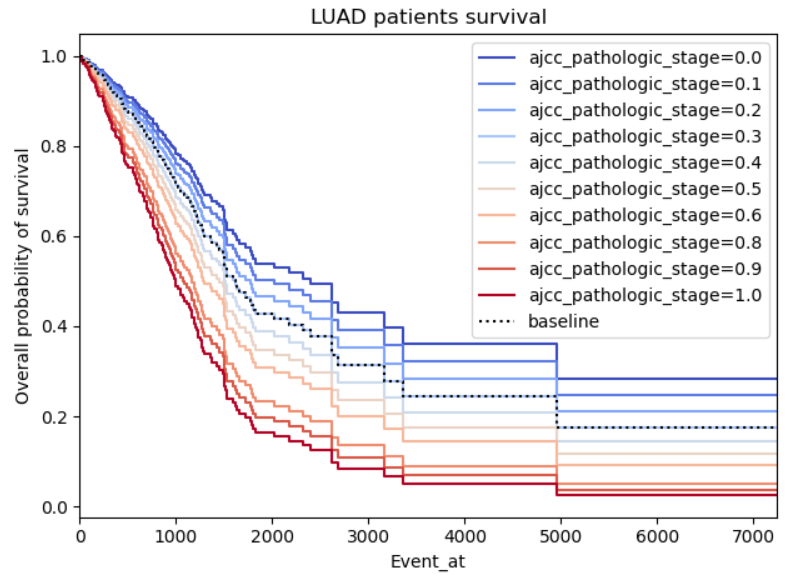
Example 1:
This one I'm providing as it looks realistic to me, there is a clear difference in the effect of the covariate when positive or negative.
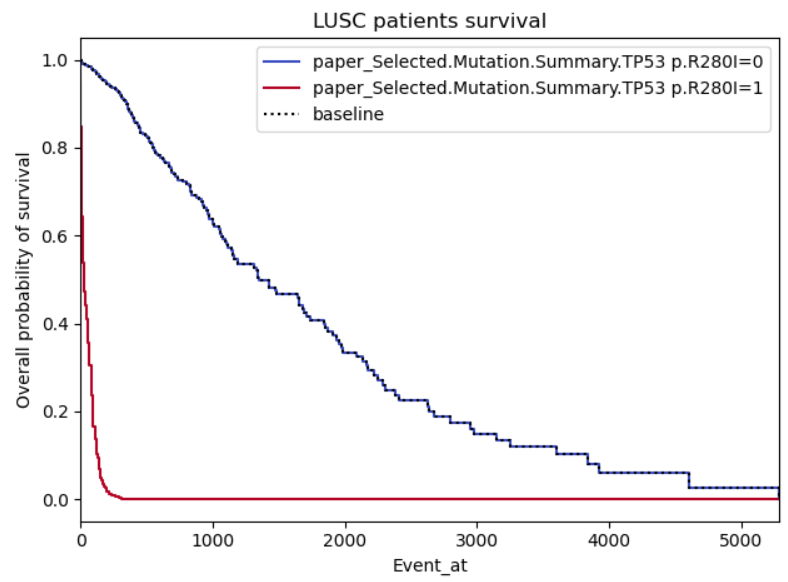
Example 2:
This one just looks gimmicky, my concern is firstly there seems to be a perfect relation between the pathologic stage of cancer and the survival, it's logical that higher stages should impact the survival negatively, however, I don't understand why the graph is so symmetric as the survivability of cancer shouldn't be proportional to the patient cancer staging.
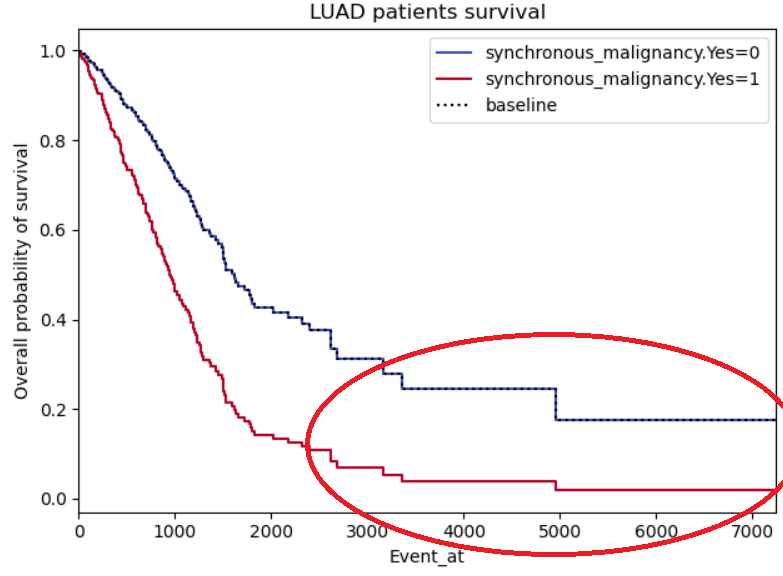
Example 3:
Same problems as for example 2, this time with a binary not categorical variable (so I'm less reluctant on disbelieving it), however, the curves do seem to symmetric, almost like it's the same curve but with a standard deviation. I also highlighted a region of interest which I don't understand why both curves stabilize (maybe lack of patients in those regions?)