Hi Charles G,
You could use the Bioconductor package karyoploteR for this. It has a function (kpHeatmap) for plots like this.
In this example I'm just using random numbers, but you can read the bed file with
gc.cont <- toGRanges("my_bed_file.bed")
To plot it you need to call plotKaryotype and specify your genome (karyoploteR can work with any other genome, including custom ones) and what part of the genome you want to plot, in this case the whole chromosome 1. After that, you'll have an empty genome plot where you can add new data, for example with kpHeatmap.
kp <- plotKaryotype(genome = "hg19", chromosomes = "chr1")
kpHeatmap(kp,data=gc.cont, y=gc.cont$gc, col=c("blue", "white", "red"))
and you'd get something like this

However, with a bit more tweaking or using other plotting functions you can represent the same values in other ways
kp <- plotKaryotype(genome = "hg19", chromosomes="chr1", main="GC content in Chromosome 1")
kpAddLabels(kp, labels = "Combined", r0=autotrack(1,4), label.margin = 0.04)
kpAxis(kp, r0=autotrack(1,4), cex=0.8)
kpBars(kp,data=gc.cont, y0=0.5, y1 = gc.cont$gc, r0=autotrack(1,4), col=colByValue(gc.cont$gc, colors = c("blue", "white", "red")), border=NA)
kpAddLabels(kp, labels = "Heatmap", r0=autotrack(2,4), label.margin = 0.04)
kpHeatmap(kp,data=gc.cont, y=gc.cont$gc, col=c("blue", "white", "red"),r0=autotrack(2,4))
kpAddLabels(kp, labels = "Bars", r0=autotrack(3,4), label.margin = 0.04)
kpAxis(kp, r0=autotrack(3,4), cex=0.8)
kpBars(kp,data=gc.cont, y0=0, y1 = gc.cont$gc, r0=autotrack(3,4), col="blue", border=NA)
kpAddLabels(kp, labels = "Line", r0=autotrack(4,4), label.margin = 0.04)
kpAxis(kp, r0=autotrack(4,4), cex=0.8)
kpLines(kp,data=gc.cont, y=gc.cont$gc, r0=autotrack(4,4))
and get an image like this one
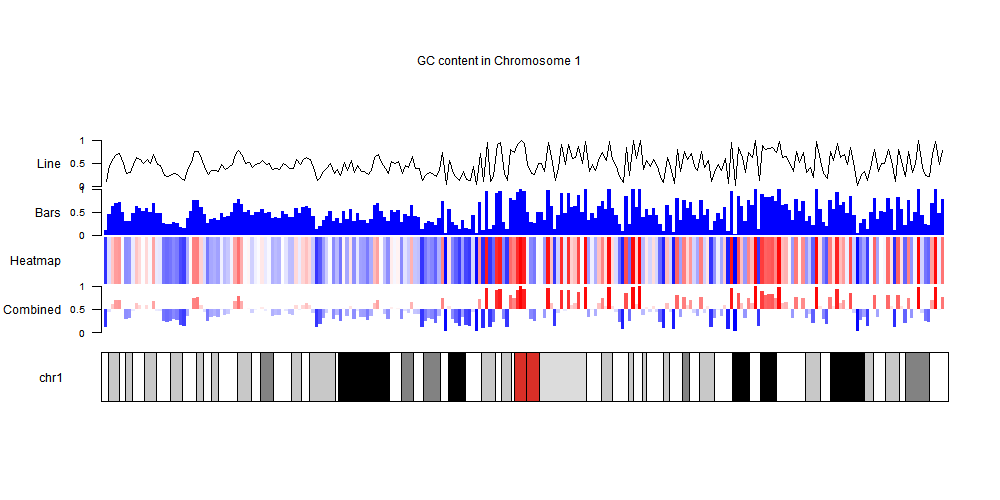
You can get more information on how to the package in the karyoploteR tutorial and the complete code for this answer on github.




You can try Gviz or ggbio which both allow heatmaps over genomic positions. The case could be made for using a circos plot for this too, with the circlize library.
thank you very much