I'm reading the preprint of a eQTM study based on data from The Framingham Heart Study (https://www.medrxiv.org/content/10.1101/2022.04.13.22273839v1.article-info). It states that the main results are available through NCBI molecular QTL browser (https://preview.ncbi.nlm.nih.gov/gap/eqtl/studies/). However, when I visited the URL, what I got is just a generic page for dbGaP.
This is very different from the screenshots shown in manuscripts mentioning the browser (For example, see https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-1142-6/figures/4). 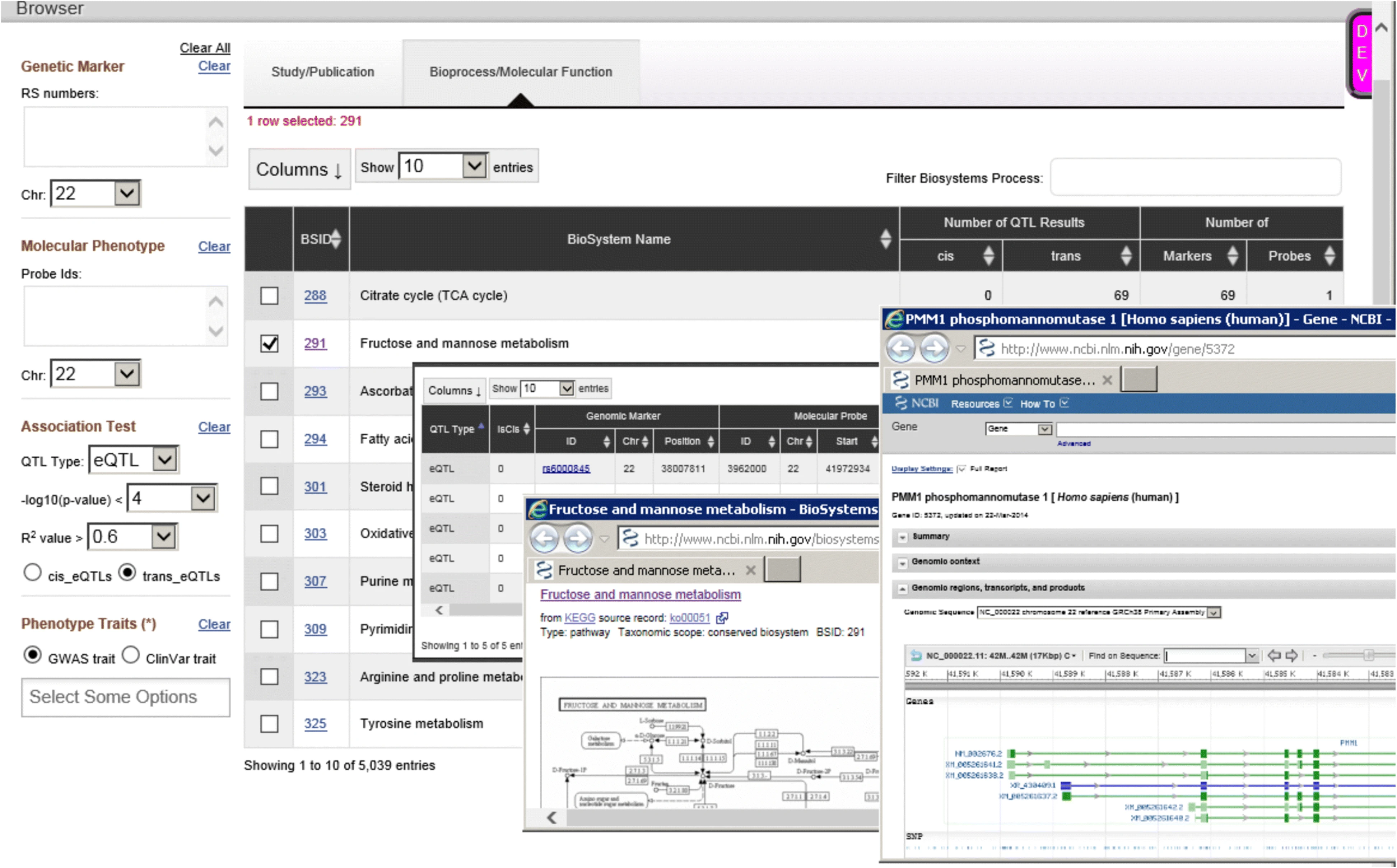



The original links are pointing to a
previewbrowser that NCBI was hosting when this paper was originally published (that is also evident from the screenshots you posted from genomebiology paper which appear to show screenshots using Microsoft Explorer). That browser is no longer available but the data you are looking for should be available from one of these links: https://www.ncbi.nlm.nih.gov/gap/advanced_search/?TERM=framingham%20heartI see. I thought the recent preprint pointing to the same URL would mean the browser is always there. Can't find the results in you link either. Contacting the corresponding author of the preprint to see what happen.