Hi!
I'm looking for a tool that can convert the 3D protein structure of a custom PDB file (not in any databases) into a 2D representation. So that the protein is laid out flat and "opened up" so you can see all loops, beta sheets and helices.
Is there any such software out there?
Thanks!

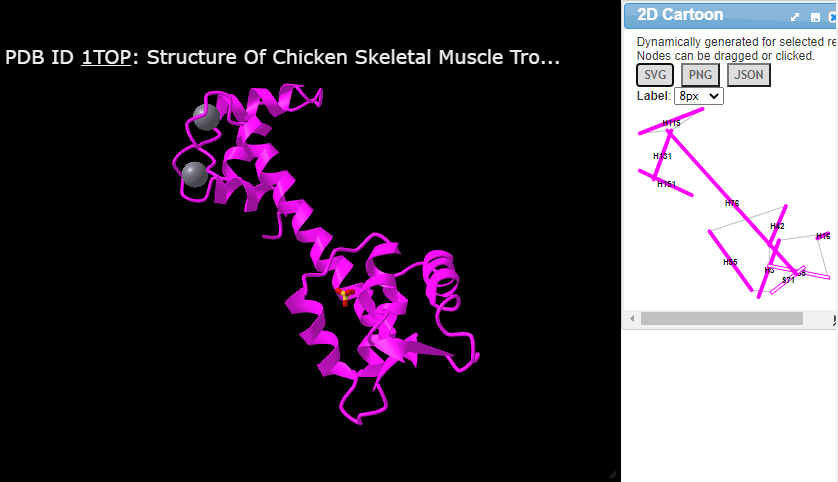

Hi Wayne! Thanks for your elaborate response. I have now looked at the tools and it seems that they're all based on existing proteins. I failed to mention (corrected now), that this is for a custom protein (my own). Hence, it is not in any databases yet. Any such tools for that purpose?
Yes, two of the three mention uploading your own protein. (Although, I'm not confident ProLego server actually works now. It was supposed to do that functionality at one time.) PDDsum let's you use 'Generate' on the server. Or you can even download it. I put that in the post already & for further emphasis I just highlighted some of the appropriate parts.
Ah, you are right! Thank you.
is it possible that you are still in this community? I used PDBSum1, it gives the different second structures compared to what chimera computed. And PTG and ProLEGO are both not work, can you tell me what software you used at last. Thanks a lot!
I think that gets into more into some sort of 'quality' assessment when you say it doesn't match a particular resource. For example, the basis for the calculations used and settings underlying could be a factor here. Most use DSSP as I understand. For example, that seem to be what Chimera is using, see here. It may indeed also factory in specifically what version of DSSP is in use, see [here](https://github.com/PDB-REDO/dssp.
Importantly, your addition here should probably be another question under its own topic. One thing you could also make clear there is the scope of your question or how it differs from here? What is your real goal? And how do the solutions posted in this thread address that or not? For example, does iCn3D work. And how is using Chimera not meeting your needs if you say that is better than PDBSum?
Importantly, this topic, as I understood it, was about generating wiring diagrams. Yours sounds like it may just be about secondary structure more generally? If you just want secondary structure assignments and not images, there's a lot more options.