Dear community members,
We are pleased to present myBrain-Seq, an open source tool for miRNA-seq data analysis of neuropsychiatric data. The source code and complete manual is available in GitHub.
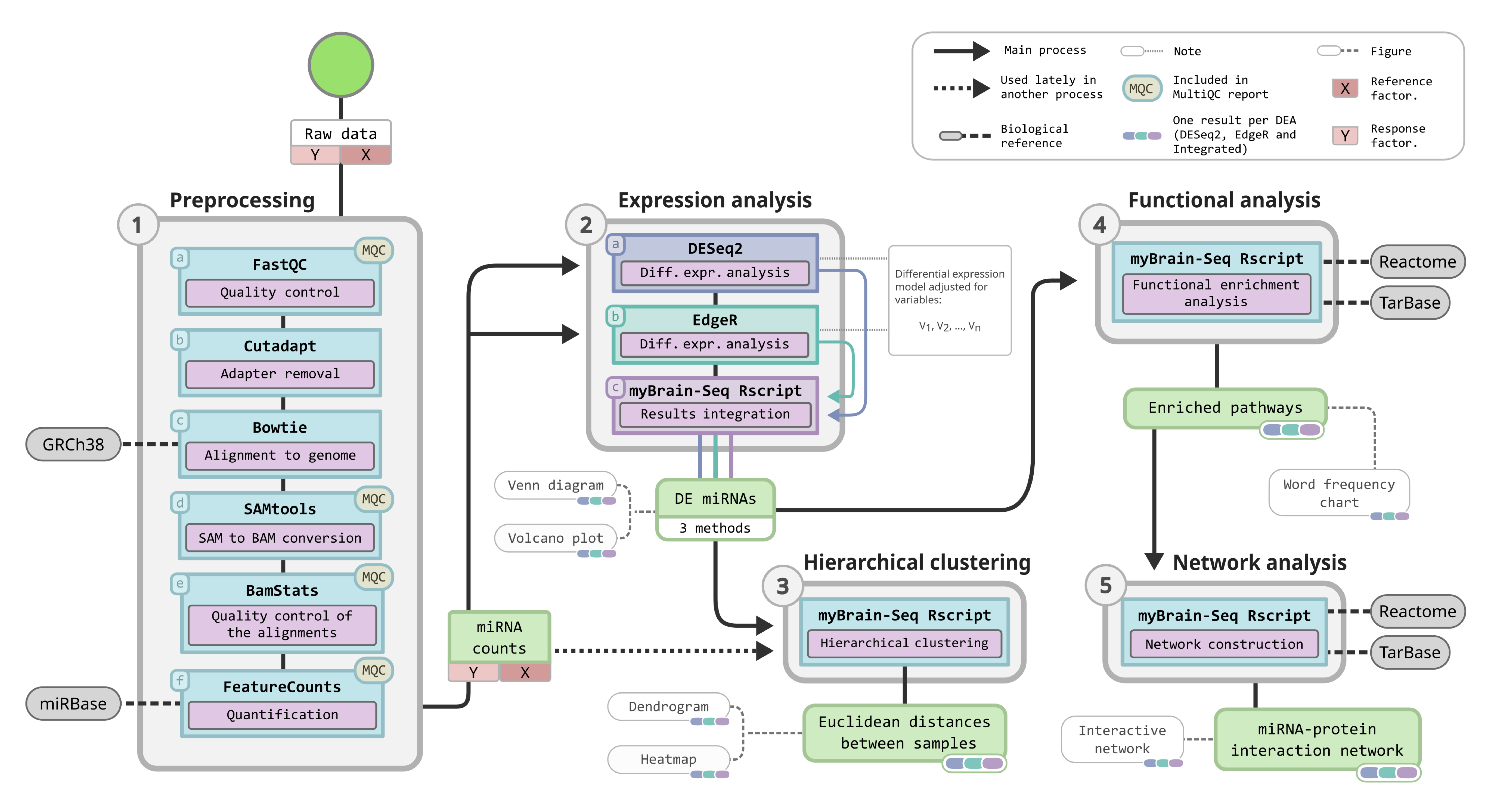
MyBrain-Seq enables simple and highly reproducible analysis of miRNA data. It incorporates commonly used processes and statistical corrections to adapt to the needs of neuropsychiatric research. In addition, it guides the researcher to obtain interpretable and publishable results, including hierarchical clustering of samples, functional analysis, network analysis, and the generation of graphics, reports, and tables. This reduces the need to resort to external solutions while ensuring traceability and reproducibility of the analytical process.
Some of the most distinguishing features of myBrain-Seq include: generation of a list of differentially expressed miRNAs from the integration of a double differential expression analysis; possibility of correcting the differential expression model for confusion variables (typically necessary in psychiatric patient data); and performance of a functional enrichment analysis corrected for assignment bias. An earlier version of myBrain-Seq has been used to support the workflow provided in the publication: MiRNA Differences Related to Treatment-Resistant Schizophrenia.
Below is a list with all the functions of myBrain-Seq v1.1:
Preprocessing
- Quality control with FastQC
- Adapter removal with Cutadapt
- Alignment with Bowtie 1
- Quality control of the alignments with FeatureCounts
Expression analysis
- Differential expression analysis with DESeq2
- Differential expression analysis with EdgeR
- Integration of both, DESeq2 and EdgeR results.
Hierarchical clustering (with several methods for computing distances)
Functional enrichment analysis (corrected for assignment bias)
Network analysis (generation of a gene-miRNA network, compatible with Cytoscape)
And here an example of the ready-to-publish graphs generated by myBrain-Seq:
You can download and start using myBrain-Seq with just one command (Docker installation needed):
docker run --rm -it -v /var/run/docker.sock:/var/run/docker.sock -v /tmp:/tmp singgroup/my-brain-seq visual_console.sh
In case you encounter any bug or you want to ask for new operations or features, please feel free to open an issue at the GitHub repository.


