I have a RNAseq dataset where I'm interested in seeing if a target gene genetically modified is upregulated (one-sided hypothesis), but at the same time, I want to assess the possible off-targets (also one-sided). I'm trying to formulate this through a Bayesian perspective for which I can provide priors to my expected target gene and predicted off-targets (based on scores from another tool independent of gene expression levels).
So, for every gene in the dataset, I will have something like this:
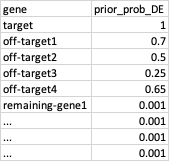
In a bayesian regression setting, I think the priors would be the log2FC, like 10, 2, 2, 0, ...
Is there a way to implement this with common workflows, like DESeq2, limma, or EdgeR? I'm open to new suggestions. Many thanks


Thank you, Gordon. This is good enough!