Somehow I am totally confused by the definition of #1 read, #2 read and the library types.
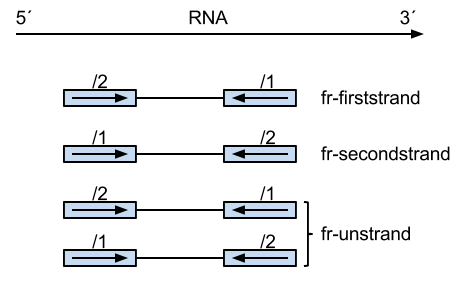
On the picture below, how /2 or /1 is defined (though it is PE)? What does it mean "the first read"? I though that the leftmost read is always the first and the right most in always the second read. Or does it mean the first read is sequenced?
+(1).png)
The definition of fr- firststrand is "enforce the rule that the right-most end of the fragment (in transcript coordinates) is the first sequenced (or only sequenced for single-end reads)".
fr- firststrand corresponds to dUTP method, where the first read sequenced matches the dUTP strand (to original RNA) but why is it considered to be the right-most end? Why not the left-most end?


IIlumina preserves the original RNA sequence, that means, that the first sequenced read is complementary to the original RNA sequence, therefore the left-most read on the figure is labeled with /1. Ist it correct? Or why is the left-most read labeled with /1?
I do not understand the labeling of the reads in the figure and why "right-most end of the fragment (in transcript coordinates) is the first sequenced" defines the fr-firststrand.
I think you need to find and revise the protocols for making the stranded libraries to notice what of the two strands is preserved af the time of making the libraries, and after analyzing for a while, you will figure out yourself the answer
I did study it and I still did not figure it out That's why I posted my question here describing exactly what I do not understand.
I am confused, the dUTP method should preserve the first strand and destroy the second strand.