Struggling with making nice plots from various NGS datasets (overlay, averaging replicates ect.), we developed a little tool to help. I would appreciate any feedback! Here a brief description:
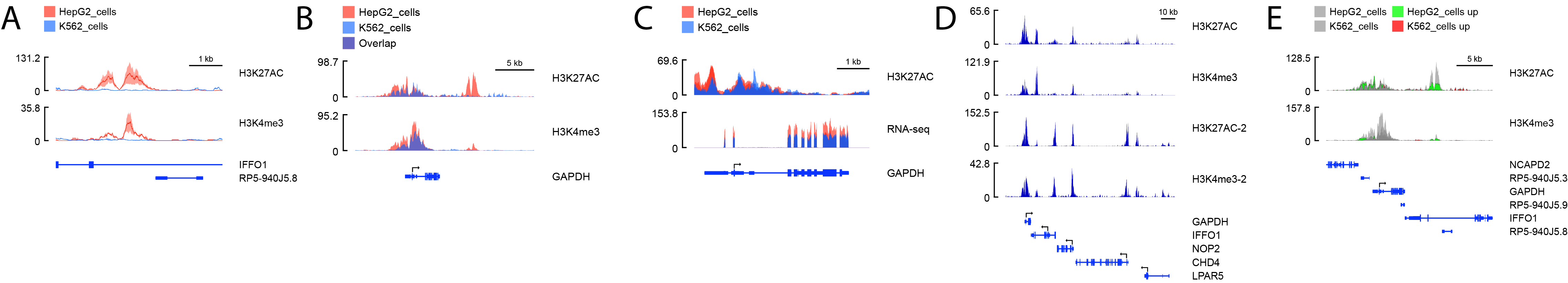
While there are sophisticated resources available for displaying NGS data, including the Integrative Genomics Viewer (IGV) and the UCSC genome browser, exporting regions and assembling figures for publication remains challenging. In particular, customizing track appearance and overlaying track replicates is a manual and time-consuming process. SparK is a novel tool which auto-generates publication-ready, high-resolution, true vector graphic figures from any NGS-based tracks, including RNA-seq, ChIP-seq, and ATAC-seq. Novel functions of SparK include averaging of replicates, plotting standard deviation tracks, and highlighting significantly changed areas. SparK is written in Python 3, making it executable on any major OS platform. Using command line prompts to generate figures allows later changes to be made very easy. For instance, if the genomic region of the plot needs to be changed, or tracks need to be added or removed, the figure can easily be re-generated within seconds without the manual process of re-exporting and re-assembling everything. After plotting with SparK, changes to the output SVG vector graphic files are simple to make, including text, lines, and colors.
Biorxiv: https://www.biorxiv.org/content/10.1101/845529v1?rss=1

