Entering edit mode
3.9 years ago
alireza.khodadadi.j
▴
70
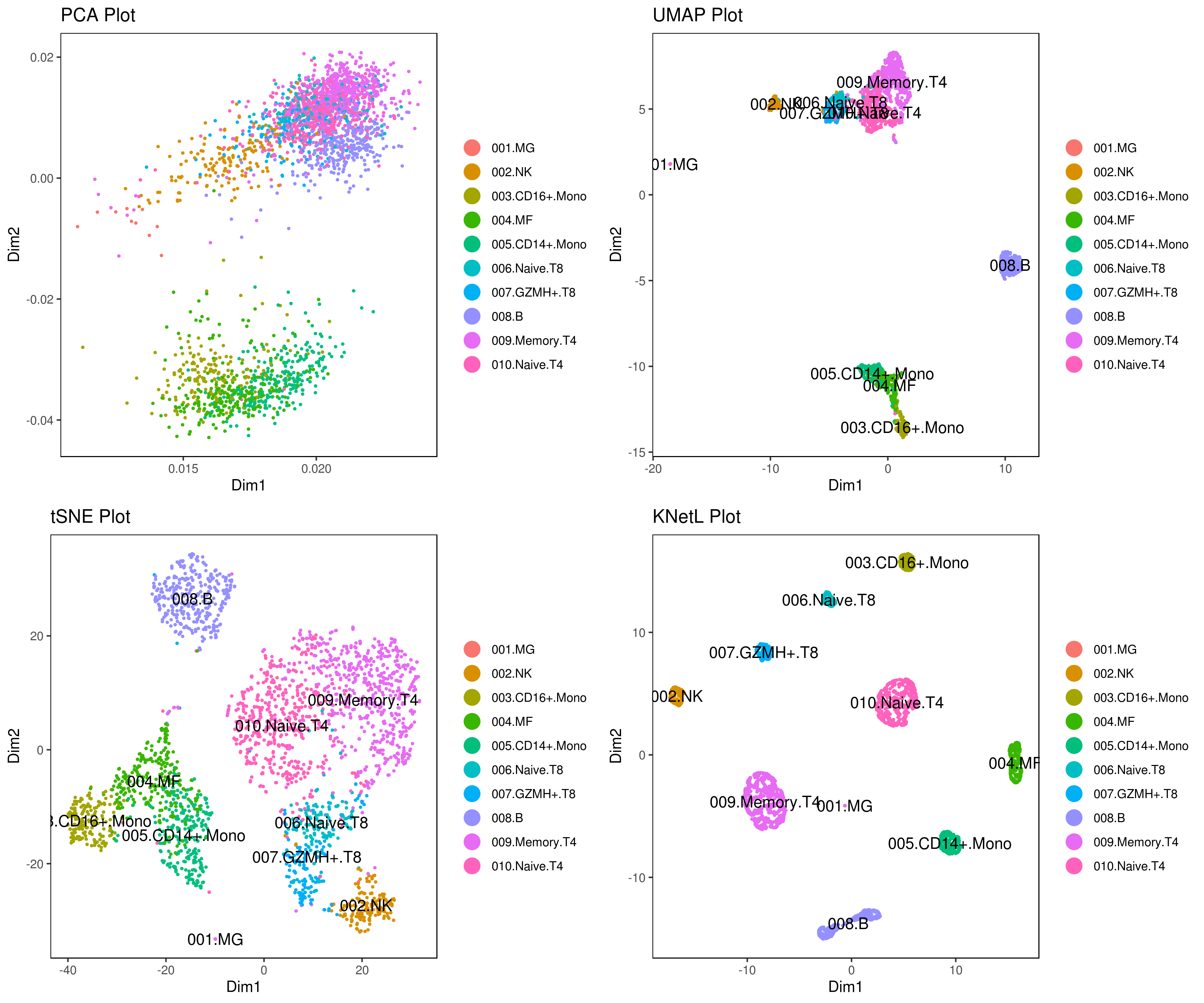
KNetL map (pronounced like "nettle") is a new dimensionality reduction developed for scRNA-Seq. It's capable of zooming and shows more detail than tSNE or UMAP.
To see more details: https://www.biorxiv.org/content/10.1101/2020.05.05.078550v1
KNetL map is available through iCellR package:
How to run KNetL, tSNE and UMAP using iCellR:
my.obj <- run.knetl(my.obj, dims = 1:20, k = 100)
my.obj <- run.pc.tsne(my.obj, dims = 1:10)
my.obj <- run.umap(my.obj, dims = 1:10)
Here we use a k value of 100, but this might not be ideal for your data.
Important Note:
- Because KNetL has a very high resolution it's best to set the dims value to 20 (this usually works best for most data) and 10 for UMPA and tSNE.
- In order to zoom, use the k value. For data with more than 5,000 cells use a k value of about 400. A k value of 400 usually works for large datasets, but you can adjust it for your intended resolution. Just like a microscope, you need to zoom in and out to see the intended amount of details.