iCn3D 2.18.0 is now available on NCBI web servers and from GitHub ( https://github.com/ncbi/icn3d ). Users can view SNPs of the 2019-nCov structures in the "Sequences & Annotations" window. Users can also access the 2D Interaction Network and 2D Interaction Map for protein-ligand or protein-protein interaction by selecting the menu "View > H-Bonds & Interactions". Another new feature is "icn3dpy", a Jupyter Notebook widget of iCn3D, which enables data scientists to use iCn3D in a Jupyter environment. Users can view a predefined display or create a custom view directly in the Jupyter Notebook. As an example the SNPs of the 2019-nCov spike (S) glycoprotein were viewed using a Jupyter Notebook and saved here as HTML.
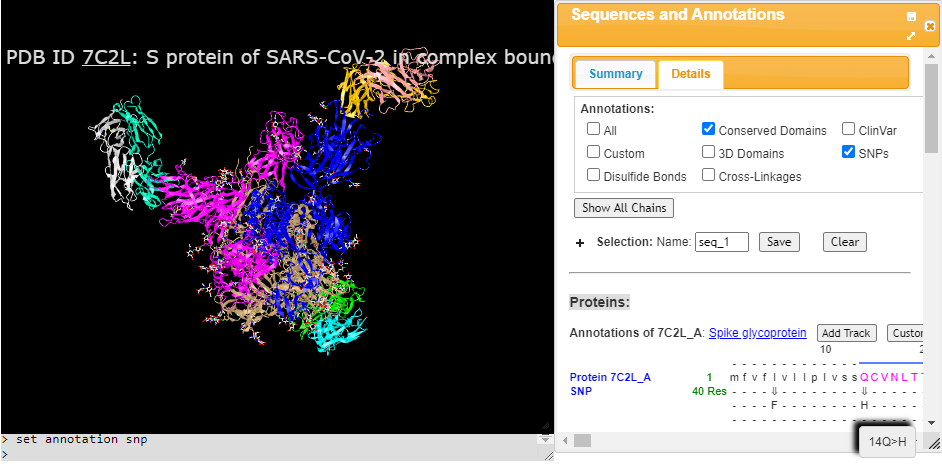
Users can now alternate wild type and mutant of SNPs in 3D, e.g., https://structure.ncbi.nlm.nih.gov/icn3d/share.html?fNpzDuUE287SBFtz8. In the "Sequences & Annotations" window, which is accessible in the menu "Analysis > Sequences & Annotations", users can click the tab "Details" and the checkbox "SNP" or "ClinVar" to see SNPs/ClinVars in the sequence. The mouseover on SNPs/ClinVars shows three buttons: "3D with scap", "Interactions", and "PDB", which allow users to alternate wild type and mutant, alternate their interactions as well, or download the coordinates. When users click "Interactions", the 2D interaction network window also pops up to show the change of interactions, which are colored in different colors.

More features are shown in the live examples: https://www.ncbi.nlm.nih.gov/Structure/icn3d/icn3d.html#gallery
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.

